Effect of oral metformin on gut microbiota characteristics and metabolite fractions in normal-weight type 2 diabetic mellitus patients
- PMID: 39257903
- PMCID: PMC11385314
- DOI: 10.3389/fendo.2024.1397034
Effect of oral metformin on gut microbiota characteristics and metabolite fractions in normal-weight type 2 diabetic mellitus patients
Abstract
Background and aims: To analyze the effect of oral metformin on changes in gut microbiota characteristics and metabolite composition in normal weight type 2 diabetic patients.
Methods: T2DM patients in the cross-sectional study were given metformin for 12 weeks. Patients with unmedicated T2DM were used as a control group to observe the metrics of T2DM patients treated with metformin regimen. 16S rDNA high-throughput gene sequencing of fecal gut microbiota of the study subjects was performed by llumina NovaSeq6000 platform. Targeted macro-metabolomics was performed on 14 cases of each of the gut microbiota metabolites of the study subjects using UPLC-MS/MS technology. Correlations between the characteristics of the gut microbiota and its metabolites, basic human parameters, glycolipid metabolism indicators, and inflammatory factors were analyzed using spearman analysis.
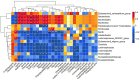
Results: Glycolipid metabolism indexes and inflammatory factors were higher in normal-weight T2DM patients than in the healthy population (P<0.05), but body weight, BMI, waist circumference, and inflammatory factor concentrations were lower in normal-weight T2DM patients than in obese T2DM patients (P<0.05). Treatment with metformin in T2DM patients improved glycolipid metabolism, but the recovery of glycolipid metabolism was more pronounced in obese T2DM patients. None of the differences in α-diversity indexes were statistically significant (P>0.05), and the differences in β-diversity were statistically significant (P <0.05). Community diversity and species richness recovered after metformin intervention compared to before, and were closer to the healthy population. We found that Anaerostipes/Xylose/Ribulose/Xylulose may play an important role in the treatment of normal-weight T2DM with metformin by improving glycemic lipids and reducing inflammation. And Metformin may play a role in obese T2DM through Romboutsia, medium-chain fatty acids (octanoic acid, decanoic acid, and dodecanoic acid).
Conclusion: Gut microbial dysbiosis and metabolic disorders were closely related to glucose-lipid metabolism and systemic inflammatory response in normal-weight T2DM patients. Metformin treatment improved glucose metabolism levels, systemic inflammation levels in T2DM patients, closer to the state of healthy population. This effect may be mediated by influencing the gut microbiota and microbial host co-metabolites, mainly associated with Anaerostipes and xylose/Ribulose/Xylulose. Metformin may exert its effects through different pathways in normal-weight versus obese T2DM patients.
Keywords: body mass index; gut microbiota; metabolomics; metformin; type 2 diabetes mellitus.
Copyright © 2024 Niu, Wang, Huang, Guo, Zhang, Sun and Jin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

