PEI-Engineered Lipid@PLGA Hybrid Nanoparticles for Multimodal Delivery of Antigens and Immune Adjuvants to the Respiratory Mucosa
- PMID: 39258393
- PMCID: PMC11670295
- DOI: 10.1002/adhm.202402688
PEI-Engineered Lipid@PLGA Hybrid Nanoparticles for Multimodal Delivery of Antigens and Immune Adjuvants to the Respiratory Mucosa
Abstract
Antigen delivery via respiratory mucosal surfaces is an interesting needle-free option for vaccination. Nonetheless, it demands for the design of especially tailored formulations. Here, lipid/poly(lactic-co-glycolic) acid (PLGA) hybrid nanoparticles (hNPs) for the combined delivery of an antigen, ovalbumin (Ova), and an adjuvant, synthetic unmethylated cytosine-phosphate-guanine oligodeoxynucleotide (CpG) motifs, is developed. A panel of Ova/CpG-loaded lipid@PLGA hNPs with tunable size and surface is attained by exploiting two lipid moieties, 1,2 distearoil-sn-glycero-3-phosphoethanolamine-poly(ethylene glycol) (DSPE-PEG) and monophosphoryl lipid A (MPLA), with or without polyethyleneimine (PEI). It is gained insights on the lipid@PLGA hNPs through a combination of techniques to analytically determine the specific moiety on the surface, the spatial distribution of the components and the internal structure of the nanoplatforms. The collected results suggest that PEI plays a role of paramount importance not only in promoting in vitro antigen escape from lysosomes and enhancing antigen cross-presentation, but also in determining the arrangement of the moieties in the final architecture of the hNPs. Though multicomponent PEI-engineered lipid@PLGA hNPs turn out as a viable strategy for delivery of antigens and adjuvant to the respiratory mucosa, tunable nanoparticle features are achievable only through the optimal selection of the components and their relative amounts.
Keywords: PLGA; antigen presentation; lipid/polymer hybrid nanoparticles; mucosal vaccination; poly(ethylenimine).
© 2024 The Author(s). Advanced Healthcare Materials published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

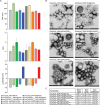
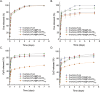
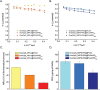
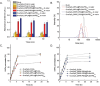


References
-
- Bessa J., Schmitz N., Hinton H. J., Schwarz K., Jegerlehner A., Bachmann M. F., Eur. J. Immunol. 2008, 38, 114. - PubMed
-
- Tsai C. J. Y., Loh J. M. S., Fujihashi K., Kiyono H., Expert Rev. Vaccines 2023, 22, 885. - PubMed
-
- Hellfritzsch S., Pharmaceutics 2019, 11, 375. - PubMed
-
- Chow M. Y.‐T., Lam J. K. W., in 2023, Respiratory Delivery of Biologics, Nucleic Acids, and Vaccines (Eds.: Lam J., Kwok P. C. L.), Springer International Publishing, Cham, 123.

