DNA double-strand break movement in heterochromatin depends on the histone acetyltransferase dGcn5
- PMID: 39258543
- PMCID: PMC11514474
- DOI: 10.1093/nar/gkae775
DNA double-strand break movement in heterochromatin depends on the histone acetyltransferase dGcn5
Abstract
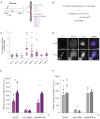
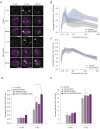
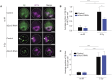
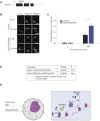
Cells employ diverse strategies to repair double-strand breaks (DSBs), a dangerous form of DNA damage that threatens genome integrity. Eukaryotic nuclei consist of different chromatin environments, each displaying distinct molecular and biophysical properties that can significantly influence the DSB-repair process. DSBs arising in the compact and silenced heterochromatin domains have been found to move to the heterochromatin periphery in mouse and Drosophila to prevent aberrant recombination events. However, it is poorly understood how chromatin components, such as histone post-translational modifications, contribute to these DSB movements within heterochromatin. Using irradiation as well as locus-specific DSB induction in Drosophila tissues and cultured cells, we find enrichment of histone H3 lysine 9 acetylation (H3K9ac) at DSBs in heterochromatin but not euchromatin. We find this increase is mediated by the histone acetyltransferase dGcn5, which rapidly localizes to heterochromatic DSBs. Moreover, we demonstrate that in the absence of dGcn5, heterochromatic DSBs display impaired recruitment of the SUMO E3 ligase Nse2/Qjt and fail to relocate to the heterochromatin periphery to complete repair. In summary, our results reveal a previously unidentified role for dGcn5 and H3K9ac in heterochromatic DSB repair and underscore the importance of differential chromatin responses at heterochromatic and euchromatic DSBs to promote safe repair.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures


References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

