CRISPR screening uncovers nucleolar RPL22 as a heterochromatin destabilizer and senescence driver
- PMID: 39258545
- PMCID: PMC11514463
- DOI: 10.1093/nar/gkae740
CRISPR screening uncovers nucleolar RPL22 as a heterochromatin destabilizer and senescence driver
Abstract
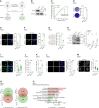
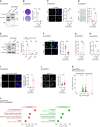
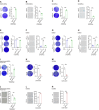
Dysfunction of the ribosome manifests during cellular senescence and contributes to tissue aging, functional decline, and development of aging-related disorders in ways that have remained enigmatic. Here, we conducted a comprehensive CRISPR-based loss-of-function (LOF) screen of ribosome-associated genes (RAGs) in human mesenchymal progenitor cells (hMPCs). Through this approach, we identified ribosomal protein L22 (RPL22) as the foremost RAG whose deficiency mitigates the effects of cellular senescence. Consequently, absence of RPL22 delays hMPCs from becoming senescent, while an excess of RPL22 accelerates the senescence process. Mechanistically, we found in senescent hMPCs, RPL22 accumulates within the nucleolus. This accumulation triggers a cascade of events, including heterochromatin decompaction with concomitant degradation of key heterochromatin proteins, specifically heterochromatin protein 1γ (HP1γ) and heterochromatin protein KRAB-associated protein 1 (KAP1). Subsequently, RPL22-dependent breakdown of heterochromatin stimulates the transcription of ribosomal RNAs (rRNAs), triggering cellular senescence. In summary, our findings unveil a novel role for nucleolar RPL22 as a destabilizer of heterochromatin and a driver of cellular senescence, shedding new light on the intricate mechanisms underlying the aging process.
© The Author(s) 2024. Published by Oxford University Press on behalf of Nucleic Acids Research.
Figures




References
MeSH terms
Substances
Grants and funding
- 2020YFA0112200/National Key Research and Development Program of China
- 81921006/National Natural Science Foundation of China
- YSBR-076/CAS Project for Young Scientists in Basic Research
- 2020YFA0804000/National Key Research and Development Program of China
- CAS-WX2022SDC-XK14/Chinese Academy of Sciences
- 2021-1045/New Cornerstone Science Foundation through the XPLORER PRIZE
- 2023092/Youth Innovation Promotion Association
- JYY2023-13/CAS Youth Interdisciplinary Team, Beijing Municipal Public Welfare Development and Reform Pilot Project for Medical Research Institutes
- 12300927/Capital Medical University
- 11000023T000002036310/Project for Technology Development of Beijing-affiliated Medical Research Institutes
- BPHR202203105/Excellent Young Talents Training Program for the Construction of Beijing Municipal University Teacher Team
- 2023IOZ0102/Initiative Scientific Research Program, Institute of Zoology, Chinese Academy of Sciences
- 2023QNRC001/CAS Special Research Assistant (SRA) Program, Young Elite Scientists Sponsorship Program by CAST
- 2023ICAC-YANFA/Key Research and Development Project of Hainan Province
- 2023ICAC-YUNXING/Science and Technology Platform Construction Project of Hainan Province
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

