Genomic evolution and rearrangement of CTX-Φ prophage elements in Vibrio cholerae during the 2018-2024 cholera outbreaks in eastern Democratic Republic of the Congo
- PMID: 39259213
- PMCID: PMC11395875
- DOI: 10.1080/22221751.2024.2399950
Genomic evolution and rearrangement of CTX-Φ prophage elements in Vibrio cholerae during the 2018-2024 cholera outbreaks in eastern Democratic Republic of the Congo
Abstract
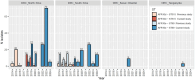
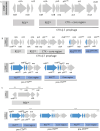
ABSTRACTBetween 2018 and 2024, we conducted systematic whole-genome sequencing and phylogenomic analysis on 263 V. cholerae O1 isolates from cholera patients across four provinces in the Democratic Republic of Congo (North-Kivu, South-Kivu, Tanganyika, and Kasai Oriental). These isolates were classified into the AFR10d and AFR10e sublineages of AFR10 lineage, originating from the third wave of the seventh El Tor cholera pandemic (7PET). Compared to the strains analysed between 2014 and 2017, both sublineages had few genetic changes in the core genome but recent isolates (2022-2024) had significant CTX prophage rearrangement. AFR10e spread across all four provinces, while AFR10d appeared to be extinct by the end of 2020. Since 2022, most V. cholerae O1 isolates exhibited significant CTX prophage rearrangements, including a tandem repeat of an environmental satellite phage RS1 downstream the ctxB toxin gene of the CTX-Φ-3 prophage on the large chromosome, as well as two or more arrayed copies of an environmental pre-CTX-Φ prophage precursor on the small chromosome. We used Illumina data for mapping and coverage estimation to identify isolates with unique CTX-Φ genomic features. Gene localization was then determined on MinION-derived assemblies, revealing an organization similar to that of non-O1 V. cholerae isolates found in Asia (O139 VC1374, and environmental O4 VCE232), but never described in V. cholerae O1 El Tor from the third wave. In conclusion, while the core genome of AFR10d and AFR10e showed minimal changes, significant alterations in the CTX-Φ and pre-CTX-Φ prophage content and organization were identified in AFR10e from 2022 onwards.
Keywords: CTX-Φ rearrangement; DRC; Vibrio cholerae; genomic evolution; prophages; whole genome.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

Similar articles
-
Genomic analysis of pathogenic isolates of Vibrio cholerae from eastern Democratic Republic of the Congo (2014-2017).PLoS Negl Trop Dis. 2020 Apr 20;14(4):e0007642. doi: 10.1371/journal.pntd.0007642. eCollection 2020 Apr. PLoS Negl Trop Dis. 2020. PMID: 32310947 Free PMC article.
-
Novel Cholera Toxin Variant and ToxT Regulon in Environmental Vibrio mimicus Isolates: Potential Resources for the Evolution of Vibrio cholerae Hybrid Strains.Appl Environ Microbiol. 2019 Jan 23;85(3):e01977-18. doi: 10.1128/AEM.01977-18. Print 2019 Feb 1. Appl Environ Microbiol. 2019. PMID: 30446560 Free PMC article.
-
Genesis of variants of Vibrio cholerae O1 biotype El Tor: role of the CTXphi array and its position in the genome.Microbiology (Reading). 2003 Jan;149(Pt 1):89-97. doi: 10.1099/mic.0.25599-0. Microbiology (Reading). 2003. PMID: 12576583
-
Whole-genome sequence comparisons reveal the evolution of Vibrio cholerae O1.Trends Microbiol. 2015 Aug;23(8):479-89. doi: 10.1016/j.tim.2015.03.010. Epub 2015 Apr 24. Trends Microbiol. 2015. PMID: 25913612 Review.
-
Evolution of new variants of Vibrio cholerae O1.Trends Microbiol. 2010 Jan;18(1):46-54. doi: 10.1016/j.tim.2009.10.003. Epub 2009 Nov 26. Trends Microbiol. 2010. PMID: 19942436 Review.
Cited by
-
Epidemiologic and Genomic Surveillance of Vibrio cholerae and Effectiveness of Single-Dose Oral Cholera Vaccine, Democratic Republic of the Congo.Emerg Infect Dis. 2025 Feb;31(2):288-297. doi: 10.3201/eid3102.241777. Emerg Infect Dis. 2025. PMID: 39983698 Free PMC article.
-
The westward spread dynamics of cholera from the eastern endemic Democratic Republic of the Congo.Emerg Microbes Infect. 2025 Dec;14(1):2437245. doi: 10.1080/22221751.2024.2437245. Epub 2024 Dec 9. Emerg Microbes Infect. 2025. PMID: 39628440 Free PMC article. No abstract available.
-
On the ability to extract MLVA profiles of Vibrio cholerae isolates from WGS data generated with Oxford Nanopore Technologies.BMC Res Notes. 2025 Jan 16;18(1):18. doi: 10.1186/s13104-025-07093-7. BMC Res Notes. 2025. PMID: 39819345 Free PMC article.
-
Effectiveness of a single dose of oral cholera vaccine: findings from epidemiological and genomic surveillance of Vibrio Cholerae in the Democratic Republic of the Congo (PICHA7 Program).medRxiv [Preprint]. 2024 Dec 16:2024.12.16.24318874. doi: 10.1101/2024.12.16.24318874. medRxiv. 2024. Update in: Emerg Infect Dis. 2025 Feb;31(2):288-297. doi: 10.3201/eid3102.241777. PMID: 39763551 Free PMC article. Updated. Preprint.
References
-
- Clemens JD, Nair GB, Ahmed T, et al. . Cholera. Lancet. 2017;390(10101):1539–1549. - PubMed
-
- Lippi D, Gotuzzo E, Caini S.. Cholera. Microbiol Spectr. 2016;4(4). - PubMed
-
- OUNOftCoH Affairs . Latin America & The Caribbean Situation Update (12-18 December 2022) OCHA; 2022. Available from: https://reliefweb.int/report/haiti/latin-america-caribbean-weekly-situat....
-
- Kanungo S, Azman AS, Ramamurthy T, et al. . Cholera. Lancet. 2022;399(10333):1429–1440. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
