Identification of TPI1 As a potential therapeutic target in pancreatic cancer with dependency of TP53 mutation using multi-omics analysis
- PMID: 39259678
- PMCID: PMC11531968
- DOI: 10.1111/cas.16302
Identification of TPI1 As a potential therapeutic target in pancreatic cancer with dependency of TP53 mutation using multi-omics analysis
Abstract
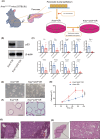
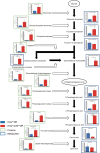
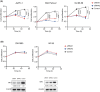
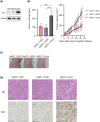
Mutations of KRAS, CDKN2A, TP53, and SMAD4 are the four major driver genes for pancreatic ductal adenocarcinoma (PDAC), of which mutations of KRAS and TP53 are the most frequently recognized. However, molecular-targeted therapies for mutations of KRAS and TP53 have not yet been developed. To identify novel molecular targets, we newly established organoids with the Kras mutation (KrasmuOR) and Trp53 loss of function using Cre transduction and CRISPR/Cas9 (Krasmu/p53muOR) from murine epithelia of the pancreatic duct in KrasLSL-G12D mice, and then analyzed the proteomic and metabolomic profiles in both organoids by mass spectrometry. Hyperfunction of the glycolysis pathway was recognized in Krasmu/p53muOR compared with KrasmuOR. Loss of function of triosephosphate isomerase (TPI1), which is involved in glycolysis, induced a reduction of cell proliferation in human PDAC cell lines with the TP53 mutation, but not in PDAC or in human fibroblasts without TP53 mutation. The TP53 mutation is clinically recognized in 70% of patients with PDAC. In the present study, protein expression of TPI1 and nuclear accumulation of p53 were recognized in the same patients with PDAC. TPI1 is a potential candidate therapeutic target for PDAC with the TP53 mutation.
Keywords: multi‐omics; organoid; p53; pancreatic cancer; triosephosphate isomerase 1 (TPI1).
© 2024 The Author(s). Cancer Science published by John Wiley & Sons Australia, Ltd on behalf of Japanese Cancer Association.
Conflict of interest statement
Honda, Kazufumi is an editorial board member of Cancer Science. The other authors have no conflict of interest.
Figures



References
-
- Kamisawa T, Wood LD, Itoi T, Takaori K. Pancreatic cancer. Lancet. 2016;388:73‐85. - PubMed
-
- Dagher OK, Schwab RD, Brookens SK, Posey AD Jr. Advances in cancer immunotherapies. Cell. 2023;186(8): P1814‐1814.e1. - PubMed
-
- Springfeld C, Jäger D, Büchler MW, et al. Chemotherapy for pancreatic cancer. Presse Med. 2019;48:E159‐E174. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

