Direct targets of MEF2C are enriched for genes associated with schizophrenia and cognitive function and are involved in neuron development and mitochondrial function
- PMID: 39259737
- PMCID: PMC11419381
- DOI: 10.1371/journal.pgen.1011093
Direct targets of MEF2C are enriched for genes associated with schizophrenia and cognitive function and are involved in neuron development and mitochondrial function
Abstract
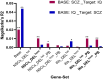
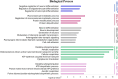
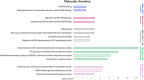
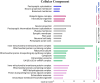
Myocyte Enhancer Factor 2C (MEF2C) is a transcription factor that plays a crucial role in neurogenesis and synapse development. Genetic studies have identified MEF2C as a gene that influences cognition and risk for neuropsychiatric disorders, including autism spectrum disorder (ASD) and schizophrenia (SCZ). Here, we investigated the involvement of MEF2C in these phenotypes using human-derived neural stem cells (NSCs) and glutamatergic induced neurons (iNs), which represented early and late neurodevelopmental stages. For these cellular models, MEF2C function had previously been disrupted, either by direct or indirect mutation, and gene expression assayed using RNA-seq. We integrated these RNA-seq data with MEF2C ChIP-seq data to identify dysregulated direct target genes of MEF2C in the NSCs and iNs models. Several MEF2C direct target gene-sets were enriched for SNP-based heritability for intelligence, educational attainment and SCZ, as well as being enriched for genes containing rare de novo mutations reported in ASD and/or developmental disorders. These gene-sets are enriched in both excitatory and inhibitory neurons in the prenatal and adult brain and are involved in a wide range of biological processes including neuron generation, differentiation and development, as well as mitochondrial function and energy production. We observed a trans expression quantitative trait locus (eQTL) effect of a single SNP at MEF2C (rs6893807, which is associated with IQ) on the expression of a target gene, BNIP3L. BNIP3L is a prioritized risk gene from the largest genome-wide association study of SCZ and has a function in mitophagy in mitochondria. Overall, our analysis reveals that either direct or indirect disruption of MEF2C dysregulates sets of genes that contain multiple alleles associated with SCZ risk and cognitive function and implicates neuron development and mitochondrial function in the etiology of these phenotypes.
Copyright: © 2024 Ali et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Li H, Radford JC, Ragusa MJ, Shea KL, McKercher SR, Zaremba JD, Soussou W, Nie Z, Kang Y-J, Nakanishi N, et al.: Transcription factor MEF2C influences neural stem/progenitor cell differentiation and maturation in vivo. Proceedings of the National Academy of Sciences 2008, 105:9397–9402. doi: 10.1073/pnas.0802876105 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

