Comparative analysis of the complete chloroplast genomes of thirteen Bougainvillea cultivars from South China with implications for their genome structures and phylogenetic relationships
- PMID: 39259741
- PMCID: PMC11389920
- DOI: 10.1371/journal.pone.0310091
Comparative analysis of the complete chloroplast genomes of thirteen Bougainvillea cultivars from South China with implications for their genome structures and phylogenetic relationships
Abstract
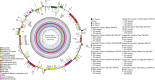
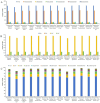
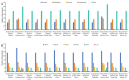
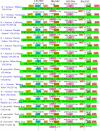
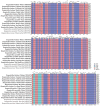
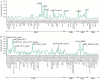
Bougainvillea spp., belonging to the Nyctaginaceae family, have high economic and horticultural value in South China. Despite the high similarity in terms of leaf appearance and hybridization among Bougainvillea species, especially Bougainvillea × buttiana, their phylogenetic relationships are very complicated and controversial. In this study, we sequenced, assembled and analyzed thirteen complete chloroplast genomes of Bougainvillea cultivars from South China, including ten B. × buttiana cultivars and three other Bougainvillea cultivars, and identified their phylogenetic relationships within the Bougainvillea genus and other species of the Nyctaginaceae family for the first time. These 13 chloroplast genomes had typical quadripartite structures, comprising a large single-copy (LSC) region (85,169-85,695 bp), a small single-copy (SSC) region (18,050-21,789 bp), and a pair of inverted-repeat (IR) regions (25,377-25,426 bp). These genomes each contained 112 different genes, including 79 protein-coding genes, 29 tRNAs and 4 rRNAs. The gene content, codon usage, simple sequence repeats (SSRs), and long repeats were essentially conserved among these 13 genomes. Single-nucleotide polymorphisms (SNPs) and insertions/deletions (indels) were detected among these 13 genomes. Four divergent regions, namely, trnH-GUG_psbA, trnS-GCU_trnG-UCC-exon1, trnS-GGA_rps4, and ccsA_ndhD, were identified from the comparative analysis of 16 Bougainvillea cultivar genomes. Among the 46 chloroplast genomes of the Nyctaginaceae family, nine genes, namely, rps12, rbcL, ndhF, rpoB, rpoC2, ndhI, psbT, ycf2, and ycf3, were found to be under positive selection at the amino acid site level. Phylogenetic relationships within the Bougainvillea genus and other species of the Nyctaginaceae family based on complete chloroplast genomes and protein-coding genes revealed that the Bougainvillea genus was a sister to the Belemia genus with strong support and that 35 Bougainvillea individuals were divided into 4 strongly supported clades, namely, Clades Ⅰ, Ⅱ, Ⅲ and Ⅳ. Clade Ⅰ included 6 individuals, which contained 2 cultivars, namely, B. × buttiana 'Gautama's Red' and B. spectabilis 'Flame'. Clades Ⅱ only contained Bougainvillea spinosa. Clade Ⅲ comprised 7 individuals of wild species. Clade Ⅳ included 21 individuals and contained 11 cultivars, namely, B. × buttiana 'Mahara', B. × buttiana 'California Gold', B. × buttiana 'Double Salmon', B. × buttiana 'Double Yellow', B. × buttiana 'Los Banos Beauty', B. × buttiana 'Big Chitra', B. × buttiana 'San Diego Red', B. × buttiana 'Barbara Karst', B. glabra 'White Stripe', B. spectabilis 'Splendens' and B. × buttiana 'Miss Manila' sp. 1. In conclusion, this study not only provided valuable genome resources but also helped to identify Bougainvillea cultivars and understand the chloroplast genome evolution of the Nyctaginaceae family.
Copyright: © 2024 Wu et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Holttum RE. The cultivated Bougainvilleas. Bougainvillea × buttiana, its variety and hybrids. Malay. Agri-hort. Ass. Mag. 1955; 12, 2–11.
-
- Gillis WT. Bougainvilleas of cultivation (Nyctaginaceae). Baileya. 1976; 20, 34–41.
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

