Modeling Lewy body disease with SNCA triplication iPSC-derived cortical organoids and identifying therapeutic drugs
- PMID: 39259788
- PMCID: PMC11389790
- DOI: 10.1126/sciadv.adk3700
Modeling Lewy body disease with SNCA triplication iPSC-derived cortical organoids and identifying therapeutic drugs
Abstract
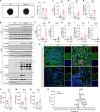
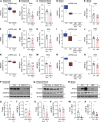
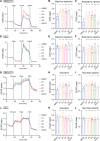
Aggregated α-synuclein (α-SYN) proteins, encoded by the SNCA gene, are hallmarks of Lewy body disease (LBD), affecting multiple brain regions. However, the specific mechanisms underlying α-SYN pathology in cortical neurons, crucial for LBD-associated dementia, remain unclear. Here, we recapitulated α-SYN pathologies in human induced pluripotent stem cells (iPSCs)-derived cortical organoids generated from patients with LBD with SNCA gene triplication. Single-cell RNA sequencing, combined with functional and molecular validation, identified synaptic and mitochondrial dysfunction in excitatory neurons exhibiting high expression of the SNCA gene, aligning with observations in the cortex of autopsy-confirmed LBD human brains. Furthermore, we screened 1280 Food and Drug Administration-approved drugs and identified four candidates (entacapone, tolcapone, phenazopyridine hydrochloride, and zalcitabine) that inhibited α-SYN seeding activity in real-time quaking-induced conversion assays with human brains, reduced α-SYN aggregation, and alleviated mitochondrial dysfunction in SNCA triplication organoids and excitatory neurons. Our findings establish human cortical LBD models and suggest potential therapeutic drugs targeting α-SYN aggregation for LBD.
Figures



References
-
- Dickson D. W., α-Synuclein and the Lewy body disorders. Curr. Opin. Neurol. 14, 423–432 (2001). - PubMed
-
- Lippa C. F., Duda J. E., Grossman M., Hurtig H. I., Aarsland D., Boeve B. F., Brooks D. J., Dickson D. W., Dubois B., Emre M., Fahn S., Farmer J. M., Galasko D., Galvin J. E., Goetz C. G., Growdon J. H., Gwinn-Hardy K. A., Hardy J., Heutink P., Iwatsubo T., Kosaka K., Lee V. M., Leverenz J. B., Masliah E., McKeith I. G., Nussbaum R. L., Olanow C. W., Ravina B. M., Singleton A. B., Tanner C. M., Trojanowski J. Q., Wszolek Z. K., DLB/PDD Working Group , DLB and PDD boundary issues: Diagnosis, treatment, molecular pathology, and biomarkers. Neurology 68, 812–819 (2007). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

