Antibiotics damage the colonic mucus barrier in a microbiota-independent manner
- PMID: 39259805
- PMCID: PMC11389797
- DOI: 10.1126/sciadv.adp4119
Antibiotics damage the colonic mucus barrier in a microbiota-independent manner
Abstract
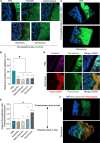
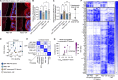
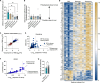
Antibiotic use is a risk factor for development of inflammatory bowel diseases (IBDs). IBDs are characterized by a damaged mucus layer, which does not separate the intestinal epithelium from the microbiota. Here, we hypothesized that antibiotics affect the integrity of the mucus barrier, which allows bacterial penetrance and predisposes to intestinal inflammation. We found that antibiotic treatment led to breakdown of the colonic mucus barrier and penetration of bacteria into the mucus layer. Using fecal microbiota transplant, RNA sequencing followed by machine learning, ex vivo mucus secretion measurements, and antibiotic treatment of germ-free mice, we determined that antibiotics induce endoplasmic reticulum stress in the colon that inhibits colonic mucus secretion in a microbiota-independent manner. This antibiotic-induced mucus secretion flaw led to penetration of bacteria into the colonic mucus layer, translocation of microbial antigens into circulation, and exacerbation of ulcerations in a mouse model of IBD. Thus, antibiotic use might predispose to intestinal inflammation by impeding mucus production.
Figures


References
-
- de Nies L., Kobras C. M., Stracy M., Antibiotic-induced collateral damage to the microbiota and associated infections. Nat. Rev. Microbiol. 21, 789–804 (2023). - PubMed
-
- Yadav S., Shah D., Dalai P., Agrawal-Rajput R., The tale of antibiotics beyond antimicrobials: Expanding horizons. Cytokine 169, 156285 (2023). - PubMed
-
- Colaço H. G., Barros A., Neves-Costa A., Seixas E., Pedroso D., Velho T., Willmann K. L., Faisca P., Grabmann G., Yi H. S., Shong M., Benes V., Weis S., Köcher T., Moita L. F., Tetracycline antibiotics induce host-dependent disease tolerance to infection. Immunity 54, 53–67.e7 (2021). - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

