HIF-2α drives hepatic Kupffer cell death and proinflammatory recruited macrophage activation in nonalcoholic steatohepatitis
- PMID: 39259813
- PMCID: PMC11665927
- DOI: 10.1126/scitranslmed.adi0284
HIF-2α drives hepatic Kupffer cell death and proinflammatory recruited macrophage activation in nonalcoholic steatohepatitis
Abstract
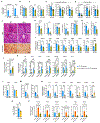
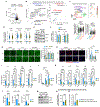
Proinflammatory hepatic macrophage activation plays a key role in the development of nonalcoholic steatohepatitis (NASH). This involves increased embryonic hepatic Kupffer cell (KC) death, facilitating the replacement of KCs with bone marrow-derived recruited hepatic macrophages (RHMs) that highly express proinflammatory genes. Moreover, phago/efferocytic activity of KCs is diminished in NASH, enhancing liver inflammation. However, the molecular mechanisms underlying these changes in KCs are not known. Here, we show that hypoxia-inducible factor 2α (HIF-2α) mediates NASH-associated decreased KC growth and efferocytosis by enhancing lysosomal stress. At the molecular level, HIF-2α stimulated mammalian target of rapamycin (mTOR)- and extracellular signal-regulated kinase-dependent inhibitory transcription factor EB (TFEB) phosphorylation, leading to decreased lysosomal and phagocytic gene expression. With increased metabolic stress and phago/efferocytic burden in NASH, these changes were sufficient to increase lysosomal stress, causing decreased efferocytosis and lysosomal cell death. Of interest, HIF-2α-dependent TFEB regulation only occurred in KCs but not RHMs. Instead, in RHMs, HIF-2α promoted mitochondrial reactive oxygen species production and proinflammatory activation by increasing ANT2 expression and mitochondrial permeability transition. Consequently, myeloid lineage-specific or KC-specific HIF-2α depletion or the inhibition of mTOR-dependent TFEB inhibition using antisense oligonucleotide treatment protected against the development of NASH in mice. Moreover, treatment with an HIF-2α-specific inhibitor reduced inflammatory and fibrogenic gene expression in human liver spheroids cultured under a NASH-like condition. Together, our results suggest that macrophage subtype-specific effects of HIF-2α collectively contribute to the proinflammatory activation of liver macrophages, leading to the development of NASH.
Figures




References
-
- Younossi ZM, Golabi P, de Avila L, Paik JM, Srishord M, Fukui N, Qiu Y, Burns L, Afendy A, Nader F, The global epidemiology of NAFLD and NASH in patients with type 2 diabetes: A systematic review and meta-analysis. J Hepatol 71, 793–801 (2019). - PubMed
-
- Loomba R, Friedman SL, Shulman GI, Mechanisms and disease consequences of nonalcoholic fatty liver disease. Cell 184, 2537–2564 (2021). - PubMed
-
- Hardy T, Oakley F, Anstee QM, Day CP, Nonalcoholic Fatty Liver Disease: Pathogenesis and Disease Spectrum. Annu Rev Pathol 11, 451–496 (2016). - PubMed
-
- Hagstrom H, Nasr P, Ekstedt M, Hammar U, Stal P, Hultcrantz R, Kechagias S, Fibrosis stage but not NASH predicts mortality and time to development of severe liver disease in biopsy-proven NAFLD. J Hepatol 67, 1265–1273 (2017). - PubMed
Publication types
MeSH terms
Substances
Grants and funding
- U01 DK130190/DK/NIDDK NIH HHS/United States
- P30 NS047101/NS/NINDS NIH HHS/United States
- UL1 TR001442/TR/NCATS NIH HHS/United States
- P42 ES010337/ES/NIEHS NIH HHS/United States
- P30 DK120515/DK/NIDDK NIH HHS/United States
- U01 AA029019/AA/NIAAA NIH HHS/United States
- R01 DK091183/DK/NIDDK NIH HHS/United States
- R01 DK099205/DK/NIDDK NIH HHS/United States
- P30 CA023100/CA/NCI NIH HHS/United States
- P50 AA011999/AA/NIAAA NIH HHS/United States
- P01 HL147835/HL/NHLBI NIH HHS/United States
- R01 DK121378/DK/NIDDK NIH HHS/United States
- R01 HL142214/HL/NHLBI NIH HHS/United States
- R01 DK106419/DK/NIDDK NIH HHS/United States
- R01 AA028550/AA/NIAAA NIH HHS/United States
- R01 DK124318/DK/NIDDK NIH HHS/United States
- U01 DK061734/DK/NIDDK NIH HHS/United States
- R01 DK124298/DK/NIDDK NIH HHS/United States
- R01 DK101737/DK/NIDDK NIH HHS/United States
- R44 DK115242/DK/NIDDK NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

