Role of ammonia-lyases in the synthesis of the dithiomethylamine ligand during [FeFe]-hydrogenase maturation
- PMID: 39260698
- PMCID: PMC11736057
- DOI: 10.1016/j.jbc.2024.107760
Role of ammonia-lyases in the synthesis of the dithiomethylamine ligand during [FeFe]-hydrogenase maturation
Abstract
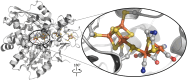
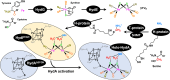
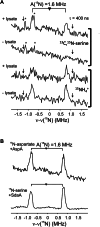
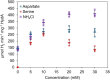
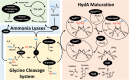
The generation of an active [FeFe]-hydrogenase requires the synthesis of a complex metal center, the H-cluster, by three dedicated maturases: the radical S-adenosyl-l-methionine (SAM) enzymes HydE and HydG, and the GTPase HydF. A key step of [FeFe]-hydrogenase maturation is the synthesis of the dithiomethylamine (DTMA) bridging ligand, a process recently shown to involve the aminomethyl-lipoyl-H-protein from the glycine cleavage system, whose methylamine group originates from serine and ammonium. Here we use functional assays together with electron paramagnetic resonance and electron-nuclear double resonance spectroscopies to show that serine or aspartate together with their respective ammonia-lyase enzymes can provide the nitrogen for DTMA biosynthesis during in vitro [FeFe]-hydrogenase maturation. We also report bioinformatic analysis of the hyd operon, revealing a strong association with genes encoding ammonia-lyases, suggesting important biochemical and metabolic connections. Together, our results provide evidence that ammonia-lyases play an important role in [FeFe]-hydrogenase maturation by delivering the ammonium required for dithiomethylamine ligand synthesis.
Keywords: GTPase; HydF; [FeFe]-hydrogenase; ammonia lyase; ammonium; dithiomethylamine; glycine cleavage system; hyd operon.
Copyright © 2024 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflicts of interest The authors declare that they have no conflicts of interest with the contents of this article.
Figures


References
-
- Lubitz W., Ogata H., Rudiger O., Reijerse E. Hydrogenases. Chem. Rev. 2014;114:4081–4148. - PubMed
-
- Kleinhaus J.T., Wittkamp F., Yadav S., Siegmund D., Apfel U.P. [FeFe]-Hydrogenases: maturation and reactivity of enzymatic systems and overview of biomimetic models. Chem. Soc. Rev. 2021;50:1668–1784. - PubMed
-
- Land H., Senger M., Berggren G., Stripp S. Current state of [FeFe]-Hydrogenase research: biodiversity and spectroscopic investigations. ACS Catal. 2020;10:7069–7086.
-
- Evans R.M., Siritanaratkul B., Megarity C.F., Pandey K., Esterle T.F., Badiani S., et al. The value of enzymes in solar fuels research - efficient electrocatalysts through evolution. Chem. Soc. Rev. 2019;48:2039–2052. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

