Atp13a5 Marker Reveals Pericyte Specification in the Mouse Central Nervous System
- PMID: 39261008
- PMCID: PMC11502228
- DOI: 10.1523/JNEUROSCI.0727-24.2024
Atp13a5 Marker Reveals Pericyte Specification in the Mouse Central Nervous System
Abstract
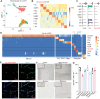
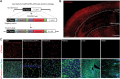
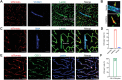
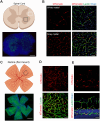
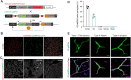
Perivascular mural cells including vascular smooth cells (VSMCs) and pericytes are integral components of the vascular system. In the central nervous system (CNS), pericytes are also indispensable for the blood-brain barrier (BBB), blood-spinal cord barrier, and blood-retinal barrier and play key roles in maintaining cerebrovascular and neuronal functions. However, the functional specifications of pericytes between CNS and peripheral organs have not been resolved at the genetic and molecular levels. Hence, the generation of reliable CNS pericyte-specific models and genetic tools remains very challenging. Here, we report a new CNS pericyte marker in mice. This putative cation-transporting ATPase 13A5 (Atp13a5) marker was identified through single-cell transcriptomics, based on its specificity to brain pericytes. We further generated a knock-in model with both tdTomato reporter and Cre recombinase. Using this model to trace the distribution of Atp13a5-positive pericytes in mice, we found that the tdTomato reporter reliably labels the CNS pericytes, including the ones in spinal cord and retina but not peripheral organs. Interestingly, brain pericytes are likely shaped by the developing neural environment, as Atp13a5-positive pericytes start to appear around murine embryonic day 15 (E15) and expand along the cerebrovasculature. Thus, Atp13a5 is a specific marker of CNS pericyte lineage, and this Atp13a5-based model is a reliable tool to explore the heterogeneity of pericytes and BBB functions in health and diseases.
Keywords: blood–brain barrier; development; mouse model; mural cells; pericyte; single-cell transcriptomics.
Copyright © 2024 Guo et al.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
