The aryl hydrocarbon receptor and FOS mediate cytotoxicity induced by Acinetobacter baumannii
- PMID: 39261458
- PMCID: PMC11390868
- DOI: 10.1038/s41467-024-52118-7
The aryl hydrocarbon receptor and FOS mediate cytotoxicity induced by Acinetobacter baumannii
Abstract
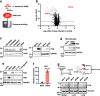
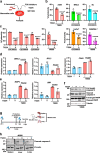
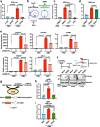
Acinetobacter baumannii is a pathogenic and multidrug-resistant Gram-negative bacterium that causes severe nosocomial infections. To better understand the mechanism of pathogenesis, we compare the proteomes of uninfected and infected human cells, revealing that transcription factor FOS is the host protein most strongly induced by A. baumannii infection. Pharmacological inhibition of FOS reduces the cytotoxicity of A. baumannii in cell-based models, and similar results are also observed in a mouse infection model. A. baumannii outer membrane vesicles (OMVs) are shown to activate the aryl hydrocarbon receptor (AHR) of host cells by inducing the host enzyme tryptophan-2,3-dioxygenase (TDO), producing the ligand kynurenine, which binds AHR. Following ligand binding, AHR is a direct transcriptional activator of the FOS gene. We propose that A. baumannii infection impacts the host tryptophan metabolism and promotes AHR- and FOS-mediated cytotoxicity of infected cells.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

