Histones and histone variant families in prokaryotes
- PMID: 39261503
- PMCID: PMC11390915
- DOI: 10.1038/s41467-024-52337-y
Histones and histone variant families in prokaryotes
Abstract
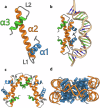
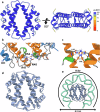
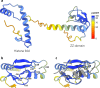
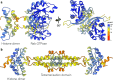
Histones are important chromatin-organizing proteins in eukaryotes and archaea. They form superhelical structures around which DNA is wrapped. Recent studies have shown that some archaea and bacteria contain alternative histones that exhibit different DNA binding properties, in addition to highly divergent sequences. However, the vast majority of these histones are identified in metagenomes and thus are difficult to study in vivo. The recent revolutionary breakthroughs in computational protein structure prediction by AlphaFold2 and RoseTTAfold allow for unprecedented insights into the potential function and structure of previously uncharacterized proteins. Here, we categorize the prokaryotic histone space into 17 distinct groups based on AlphaFold2 predictions. We identify a superfamily of histones, termed α3 histones, which are common in archaea and present in several bacteria. Importantly, we establish the existence of a large family of histones throughout archaea and in some bacteriophages that, instead of wrapping DNA, bridge DNA, thereby diverging from conventional nucleosomal histones.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Luijsterburg, M. S., White, M. F., Driel, R. V. & Dame, R. T. The major architects of chromatin: architectural proteins in bacteria, archaea and eukaryotes. Crit. Rev. Biochem. Mol. Biol.9238, 393–418 (2009). - PubMed
-
- Sandman, K., Krzycki, J. A., Dobrinski, B., Lurz, R. & Reeve, J. N. HMf, a DNA-binding protein isolated from the hyperthermophilic archaeon Methanothermus fervidus, is most closely related to histones. Proceedings of the National Academy of Sciences of the United States of America (1990). - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

