Correction of exon 2, exon 2-9 and exons 8-9 duplications in DMD patient myogenic cells by a single CRISPR/Cas9 system
- PMID: 39261505
- PMCID: PMC11390959
- DOI: 10.1038/s41598-024-70075-5
Correction of exon 2, exon 2-9 and exons 8-9 duplications in DMD patient myogenic cells by a single CRISPR/Cas9 system
Abstract
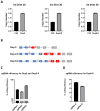
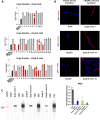
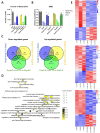
Duchenne Muscular dystrophy (DMD), a yet-incurable X-linked recessive disorder that results in muscle wasting and loss of ambulation is due to mutations in the dystrophin gene. Exonic duplications of dystrophin gene are a common type of mutations found in DMD patients. In this study, we utilized a single guide RNA CRISPR strategy targeting intronic regions to delete the extra duplicated regions in patient myogenic cells carrying duplication of exon 2, exons 2-9, and exons 8-9 in the DMD gene. Immunostaining on CRISPR-corrected derived myotubes demonstrated the rescue of dystrophin protein. Subsequent RNA sequencing of the DMD cells indicated rescue of genes of dystrophin related pathways. Examination of predicted close-match off-targets evidenced no aberrant gene editing at these loci. Here, we further demonstrate the efficiency of a single guide CRISPR strategy capable of deleting multi-exon duplications in the DMD gene without significant off target effect. Our study contributes valuable insights into the safety and efficacy of using single guide CRISPR strategy as a potential therapeutic approach for DMD patients with duplications of variable size.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures
Similar articles
-
Molecular correction of Duchenne muscular dystrophy by splice modulation and gene editing.RNA Biol. 2021 Jul;18(7):1048-1062. doi: 10.1080/15476286.2021.1874161. Epub 2021 Jan 20. RNA Biol. 2021. PMID: 33472516 Free PMC article. Review.
-
CRISPR/Cas9-based genome editing for the modification of multiple duplications that cause Duchenne muscular dystrophy.Gene Ther. 2022 Dec;29(12):730-737. doi: 10.1038/s41434-022-00336-3. Epub 2022 May 9. Gene Ther. 2022. PMID: 35534612
-
Correction of Three Prominent Mutations in Mouse and Human Models of Duchenne Muscular Dystrophy by Single-Cut Genome Editing.Mol Ther. 2020 Sep 2;28(9):2044-2055. doi: 10.1016/j.ymthe.2020.05.024. Epub 2020 May 30. Mol Ther. 2020. PMID: 32892813 Free PMC article.
-
From gRNA Identification to the Restoration of Dystrophin Expression: A Dystrophin Gene Correction Strategy for Duchenne Muscular Dystrophy Mutations Using the CRISPR-Induced Deletion Method.Methods Mol Biol. 2018;1687:267-283. doi: 10.1007/978-1-4939-7374-3_19. Methods Mol Biol. 2018. PMID: 29067670
-
Therapeutic Applications of CRISPR/Cas for Duchenne Muscular Dystrophy.Curr Gene Ther. 2017;17(4):301-308. doi: 10.2174/1566523217666171121165046. Curr Gene Ther. 2017. PMID: 29173172 Review.
Cited by
-
Targeted correction of megabase-scale CNTN6 duplication in induced pluripotent stem cells and impacts on gene expression.PeerJ. 2025 Jan 20;13:e18567. doi: 10.7717/peerj.18567. eCollection 2025. PeerJ. 2025. PMID: 39850828 Free PMC article.
-
Gene Editing for Duchenne Muscular Dystrophy: From Experimental Models to Emerging Therapies.Degener Neurol Neuromuscul Dis. 2025 Apr 12;15:17-40. doi: 10.2147/DNND.S495536. eCollection 2025. Degener Neurol Neuromuscul Dis. 2025. PMID: 40241992 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

