Comprehensive analysis of scRNA-Seq and bulk RNA-Seq reveals ubiquitin promotes pulmonary fibrosis in chronic pulmonary diseases
- PMID: 39261509
- PMCID: PMC11390722
- DOI: 10.1038/s41598-024-70659-1
Comprehensive analysis of scRNA-Seq and bulk RNA-Seq reveals ubiquitin promotes pulmonary fibrosis in chronic pulmonary diseases
Abstract
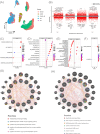
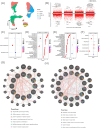
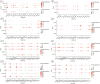
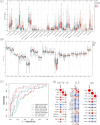
It is estimated that there are 544.9 million people suffering from chronic respiratory diseases in the world, which is the third largest chronic disease. Although there are various clinical treatment methods, there is no specific drug for chronic pulmonary diseases, including chronic obstructive pulmonary disease (COPD), interstitial lung disease (ILD) and idiopathic pulmonary fibrosis (IPF). Therefore, it is urgent to clarify the pathological mechanism and medication development. Single-cell transcriptome data of human and mouse from GEO database were integrated by "Harmony" algorithm. The data was standardized and normalized by using "Seurat" package, and "SingleR" algorithm was used for cell grouping annotation. The "Findmarker" function is used to find differentially expressed genes (DEGs), which were enriched and analyzed by using "clusterProfiler", and a protein interaction network was constructed for DEGs, and four algorithms are used to find the hub genes. The expression of hub genes were analyzed in independent human and mouse single-cell transcriptome data. Bulk RNA data were used to integrate by the "SVA" function, verify the expression levels of hub genes and build a diagnostic model. The L1000FWD platform was used to screen potential drugs. Through exploring the similarities and differences by integrated single-cell atlas, we found that the lung parenchymal cells showed abnormal oxidative stress, cell matrix adhesion and ubiquitination in COPD, corona virus disease 2019 (COVID-19), ILD and IPF. Meanwhile, the lung resident immune cells showed abnormal Toll-like receptor signals, interferon signals and ubiquitination. However, unlike acute pneumonia (COVID-19), chronic pulmonary disease shows enhanced ubiquitination. This phenomenon was confirmed in independent external human single-cell atlas, but unfortunately, it was not confirmed in mouse single-cell atlas of bleomycin-induced pulmonary fibrosis model and influenza virus-infected mouse model, which means that the model needs to be optimized. In addition, the bulk RNA-Seq data of COVID-19, ILD and IPF was integrated, and we found that the immune infiltration of lung tissue was enhanced, consistent with the single-cell level, UBA52, UBB and UBC were low expressed in COVID-19 and high expressed in ILD, and had a strong correlation with the expression of cell matrix adhesion genes. UBA52 and UBB have good diagnostic efficacy, and salermide and SSR-69071 can be used as their candidate drugs. Our study found that the disorder of protein ubiquitination in chronic pulmonary diseases is an important cause of pathological phenotype of pulmonary fibrosis by integrating scRNA-Seq and bulk RNA-Seq, which provides a new horizons for clinicopathology, diagnosis and treatment.
Keywords: Bulk RNA sequencing; Chronic pulmonary diseases; Immune infiltration; Single-cell RNA sequencing; Ubiquitination.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures







References
-
- GBD Chronic Respiratory Disease Collaborators. Prevalence and attributable health burden of chronic respiratory diseases, 1990–2017: A systematic analysis for the Global Burden of Disease Study 2017. Lancet Respir. Med.8(6), 585–596. 10.1016/S2213-2600(20)30105-3 (2020). 10.1016/S2213-2600(20)30105-3 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

