Aging is associated with functional and molecular changes in distinct hematopoietic stem cell subsets
- PMID: 39261515
- PMCID: PMC11391069
- DOI: 10.1038/s41467-024-52318-1
Aging is associated with functional and molecular changes in distinct hematopoietic stem cell subsets
Abstract
Age is a risk factor for hematologic malignancies. Attributes of the aging hematopoietic system include increased myelopoiesis, impaired adaptive immunity, and a functional decline of the hematopoietic stem cells (HSCs) that maintain hematopoiesis. Changes in the composition of diverse HSC subsets have been suggested to be responsible for age-related alterations, however, the underlying regulatory mechanisms are incompletely understood in the context of HSC heterogeneity. In this study, we investigated how distinct HSC subsets, separated by CD49b, functionally and molecularly change their behavior with age. We demonstrate that the lineage differentiation of both lymphoid-biased and myeloid-biased HSC subsets progressively shifts to a higher myeloid cellular output during aging. In parallel, we show that HSCs selectively undergo age-dependent gene expression and gene regulatory changes in a progressive manner, which is initiated already in the juvenile stage. Overall, our studies suggest that aging intrinsically alters both cellular and molecular properties of HSCs.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



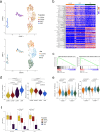

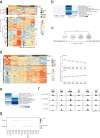
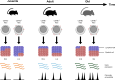
References
Publication types
MeSH terms
Grants and funding
- 2016.0131/Knut och Alice Wallenbergs Stiftelse (Knut and Alice Wallenberg Foundation)
- TJ2017-0074, PR2017-0047/Barncancerfonden (Swedish Childhood Cancer Foundation)
- CAN2017/583, 20 1062 PjF/Cancerfonden (Swedish Cancer Society)
- 19 0092 SIA, 20 1222PjF/Cancerfonden (Swedish Cancer Society)
- 2016-02331/Vetenskapsrådet (Swedish Research Council)
LinkOut - more resources
Full Text Sources
Medical

