Ancient Rapanui genomes reveal resilience and pre-European contact with the Americas
- PMID: 39261618
- PMCID: PMC11390480
- DOI: 10.1038/s41586-024-07881-4
Ancient Rapanui genomes reveal resilience and pre-European contact with the Americas
Abstract
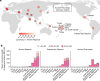
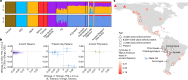
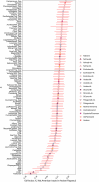
Rapa Nui (also known as Easter Island) is one of the most isolated inhabited places in the world. It has captured the imagination of many owing to its archaeological record, which includes iconic megalithic statues called moai1. Two prominent contentions have arisen from the extensive study of Rapa Nui. First, the history of the Rapanui has been presented as a warning tale of resource overexploitation that would have culminated in a major population collapse-the 'ecocide' theory2-4. Second, the possibility of trans-Pacific voyages to the Americas pre-dating European contact is still debated5-7. Here, to address these questions, we reconstructed the genomic history of the Rapanui on the basis of 15 ancient Rapanui individuals that we radiocarbon dated (1670-1950 CE) and whole-genome sequenced (0.4-25.6×). We find that these individuals are Polynesian in origin and most closely related to present-day Rapanui, a finding that will contribute to repatriation efforts. Through effective population size reconstructions and extensive population genetics simulations, we reject a scenario involving a severe population bottleneck during the 1600s, as proposed by the ecocide theory. Furthermore, the ancient and present-day Rapanui carry similar proportions of Native American admixture (about 10%). Using a Bayesian approach integrating genetic and radiocarbon dates, we estimate that this admixture event occurred about 1250-1430 CE.
© 2024. The Author(s).
Conflict of interest statement
O.D. is a current employee of the Regeneron Genetics Center, which is a subsidiary of Regeneron Pharmaceuticals. The remaining authors declare no competing interests.
Figures



References
-
- Hunt, T. L. & Lipo, C. P. The Statues That Walked: Unraveling the Mystery of Easter Island (Free Press, 2011).
-
- Diamond, J. M. Collapse: How Societies Choose to Fail or Succeed (Penguin, 2006).
-
- Bahn, P. G. & Flenley, J. Easter Island, Earth Island (Thames and Hudson, 1992).
-
- DiNapoli, R. J., Lipo, C. P. & Hunt, T. L. Revisiting warfare, monument destruction, and the ‘Huri Moai’ phase in Rapa Nui (Easter Island) culture history. J. Pac. Archaeol.12, 1–24 (2020).
-
- Moreno-Mayar, J. V. et al. Genome-wide ancestry patterns in Rapanui suggest pre-European admixture with native Americans. Curr. Biol.24, 2518–2525 (2014). - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources

