Interaction of chikungunya virus glycoproteins with macrophage factors controls virion production
- PMID: 39261662
- PMCID: PMC11480453
- DOI: 10.1038/s44318-024-00193-3
Interaction of chikungunya virus glycoproteins with macrophage factors controls virion production
Abstract
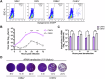
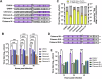
Despite their role as innate sentinels, macrophages can serve as cellular reservoirs of chikungunya virus (CHIKV), a highly-pathogenic arthropod-borne alphavirus that has caused large outbreaks among human populations. Here, with the use of viral chimeras and evolutionary selection analysis, we define CHIKV glycoproteins E1 and E2 as critical for virion production in THP-1 derived human macrophages. Through proteomic analysis and functional validation, we further identify signal peptidase complex subunit 3 (SPCS3) and eukaryotic translation initiation factor 3 subunit K (eIF3k) as E1-binding host proteins with anti-CHIKV activities. We find that E1 residue V220, which has undergone positive selection, is indispensable for CHIKV production in macrophages, as its mutation attenuates E1 interaction with the host restriction factors SPCS3 and eIF3k. Finally, we show that the antiviral activity of eIF3k is translation-independent, and that CHIKV infection promotes eIF3k translocation from the nucleus to the cytoplasm, where it associates with SPCS3. These functions of CHIKV glycoproteins late in the viral life cycle provide a new example of an intracellular evolutionary arms race with host restriction factors, as well as potential targets for therapeutic intervention.
Keywords: Alphavirus E1 Glycoprotein; Chikungunya Virus; Evolutionary Selection; Macrophage; eIF3k.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures









References
-
- Brault AC, Foy BD, Myles KM, Kelly CLH, Higgs S, Weaver SC, Olson KE, Miller BR, Powers AM (2004) Infection patterns of o’nyong nyong virus in the malaria-transmitting mosquito, Anopheles gambiae. Insect Mol Biol 13:625–635 - PubMed
-
- Bréhin A-C, Casadémont I, Frenkiel M-P, Julier C, Sakuntabhai A, Desprès P (2009) The large form of human 2′,5′-oligoadenylate synthetase (OAS3) exerts antiviral effect against chikungunya virus. Virology 384:216–222 - PubMed
MeSH terms
Substances
Grants and funding
- R35GM153408/Foundation for the National Institutes of Health (FNIH)
- AI007323/Ruth L. Kirschstein National Research Service Award
- T32 AI007323/AI/NIAID NIH HHS/United States
- Pilot Grant/UC | UCLA | Center for AIDS Research, University of California Los Angeles (CFAR)
- Sydney Finegold Post-Doctoral Fellow Award/UC | UCLA | Department of Microbiology, Immunology and Molecular Genetics, University of California Los Angeles
- R35 GM153408/GM/NIGMS NIH HHS/United States
- R01AI158704/Foundation for the National Institutes of Health (FNIH)
- HARC Collaborative development Award/HIV Accessory and Regulatory Complexes Center
- CRN-20-637544/University of California (UC)
- GM089778/Foundation for the National Institutes of Health (FNIH)
- R01 GM089778/GM/NIGMS NIH HHS/United States
- R01 AI158704/AI/NIAID NIH HHS/United States
- P30 AI028697/AI/NIAID NIH HHS/United States

