Bioinformatic prediction of miR-320a as a potential negative regulator of CDGSH iron-sulfur domain 2 (CISD2), involved in lung adenocarcinoma bone metastasis via MYC activation, and associated with tumor immune infiltration
- PMID: 39262456
- PMCID: PMC11385248
- DOI: 10.21037/tcr-24-1188
Bioinformatic prediction of miR-320a as a potential negative regulator of CDGSH iron-sulfur domain 2 (CISD2), involved in lung adenocarcinoma bone metastasis via MYC activation, and associated with tumor immune infiltration
Abstract
Background: Ferroptosis, a form of regulated cell death associated with iron-dependent lipid peroxidation, plays a role in cancer progression. However, the specific mechanisms of ferroptosis in lung adenocarcinoma (LUAD) bone metastasis (BM) remain unclear. Using bioinformatics analysis, this study sought to identify the ferroptosis-associated genes involved in BM in LUAD, thus providing potential novel targets for the treatment of BM in LUAD.
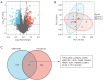
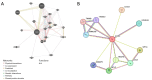
Methods: The RNA expression dataset GSE10799 was acquired from the Gene Expression Omnibus (GEO) database, and intersected with the ferroptosis dataset to identify ferroptosis-related differentially expressed genes (DEGs). The expression of candidate genes and their correlation with the prognosis of LUAD patients were validated in The Cancer Genome Atlas (TCGA) database. A protein gene interaction network was constructed using GeneMania and Retrieval of Interacting Genes/Proteins (STRING) databases. The association between the candidate genes and immune cells was assessed via TCGA and Tumor IMmune Estimation Resource (TIMER) databases. The potential mechanisms were elucidated by a gene set enrichment analysis (GSEA). The relevant microRNAs (miRNAs or miRs) that bind to the 3'untranslated region (3'UTR) end of candidate genes' mRNA was explored using the TargetScan database. The expression of these candidate miRNAs in LUAD was validated and the correlation between candidate miRNAs and candidate mRNAs was tested using the TCGA database. Finally, the clinical data of 40 LUAD patients were retrospectively analyzed to evaluate the clinical value of candidate gene expression for LUAD BM patients.
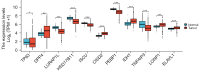
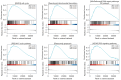
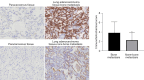
Results: In this research, 15 ferroptosis-related DEGs in LUAD BM were identified. TCGA database analysis indicated that patients with low levels of CDGSH iron-sulfur domain 2 (CISD2) in LUAD had better disease-specific survival (DSS), overall survival (OS), and a better progression-free interval (PFI) than those with high levels of CISD2. The TIMER database results show that the expression of CISD2 is correlated with the infiltration levels of various immune cells. The GSEA indicated that CISD2 might influence biological activity in LUAD by participating in cell-cycle regulation, mitochondrial translation, DNA damage repair, c-Myc (MYC) activation, and the P53 signaling pathway. Through the combined analysis of the TargetScan and TCGA databases, hsa-miR-320a was identified as the optimal upstream regulatory miRNA. The immunohistochemistry data indicated that the positive CISD2 expression rates and immunohistochemistry scores of the patients with BM were significantly higher than those of the patients without BM (P<0.05). The high expression of CISD2 is a significant risk factor for BM in LUAD.
Conclusions: The downregulation of CISD2 expression may extend DSS, OS, and the PFI of LUAD patients. Thus, CISD2 could serve as a novel predictive biomarker for LUAD patients. Further, miR-320a might negatively regulate CISD2 and participate in LUAD BM by activating MYC. These data provide a potential perspective for developing anticancer therapies for LUAD-BM patients.
Keywords: Lung adenocarcinoma (LUAD); bioinformatics analysis; bone metastasis (BM); ferroptosis; prognosis.
2024 Translational Cancer Research. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-24-1188/coif). The authors have no conflicts of interest to declare.
Figures




References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
