Unveiling the role of RARs in stomach adenocarcinoma: clinical implications and prognostic biomarkers
- PMID: 39262490
- PMCID: PMC11384927
- DOI: 10.21037/tcr-23-2154
Unveiling the role of RARs in stomach adenocarcinoma: clinical implications and prognostic biomarkers
Abstract
Background: Retinoic acid receptors (RARs) family are known to play a significant role in the occurrence and development of tumors. However, the relationship between RARs and stomach adenocarcinoma (STAD) has not yet been clearly identified. The aim of this study is to evaluate the expression profile and clinical value of the RARs family in STAD.
Methods: The expression level, clinical characteristics, prognostic value, immunity-related evaluations, genetic alteration and methylation site of RARs in STAD were explored using a series of online databases including gene expression profiling interactive analysis (GEPIA), tumor immune estimation resource (TIMER), University of Alabama at Birmingham cancer data (UALCAN), Human Protein Atlas (HPA), Kaplan-Meier plotter, gene set cancer analysis (GSCA), cBioPortal, MethSurv, GeneMANIA, LinkedOmics, Metascape, Search tool for the retrieval of interacting genes (STRING), tumor immune single-cell hub (TISCH) and cancer cell line encyclopedia (CCLE).
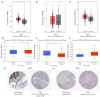
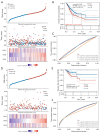
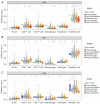
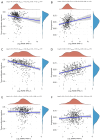
Results: We discovered dramatically increased expression of RARA and decreased expression of RARB in STAD tissues, and many clinical variables were closely related to RARs. Notably, higher expressions of RARA and RARB as well as lower expression of RARG correlated with worse overall survival (OS) for STAD patients. The clinical value of prognostic model indicated that RARs were identified to be potential prognostic biomarkers for STAD patients. Moreover, RARB was closely related to immune cell infiltration, which had effect on the role of RARB in STAD prognosis. And the genetic alteration of RARB was significantly associated with the longer disease-free survival (DFS) of STAD patients. Additionally, some CpG sites of the RARs family were related with the prognosis of STAD patients. Functional enrichment analyses indicated that several pathways in STAD might be pivotal pathways regulated by RARs. At the single-cell level, there was some extent of infiltration of tumor microenvironment-related cells in the RARs expression in STAD.
Conclusions: Our results evaluated the expression profile and clinical values of RARs in patients with STAD, which provided a basis for future in-depth exploration of the specific mechanisms of each member of RARs in STAD.
Keywords: Retinoic acid receptors family members (RARs family members); bioinformatics analysis; prognosis; stomach adenocarcinoma (STAD).
2024 Translational Cancer Research. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://tcr.amegroups.com/article/view/10.21037/tcr-23-2154/coif). The authors have no conflicts of interest to declare.
Figures






References
LinkOut - more resources
Full Text Sources
