Comparison of cell-free and small extracellular-vesicle-associated DNA by sequencing plasma of lung cancer patients
- PMID: 39262778
- PMCID: PMC11389540
- DOI: 10.1016/j.isci.2024.110742
Comparison of cell-free and small extracellular-vesicle-associated DNA by sequencing plasma of lung cancer patients
Abstract
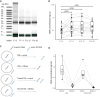
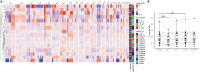
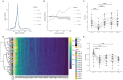
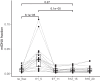
Blood contains multiple analytes that can be used as liquid biopsy to analyze cancer. Mutations have been detected in DNA associated with small extracellular vesicles (sEVs). The genome-wide composition and structure of sEV DNA remains poorly characterized, and whether sEVs are enriched in tumor signal compared to cell-free DNA (cfDNA) is unclear. Here, using whole-genome sequencing from lung cancer patients we determined that the tumor fraction and heterogeneity are comparable between DNA associated with sEV (<200 nm) and matched plasma cfDNA. sEV DNA, obtained with size-exclusion chromatography, is composed of short ∼150-180 bp fragments and long >1000 bp fragments poor in tumor signal. The structural patterns of sEV DNA are related to plasma cfDNA. Mitochondrial DNA is relatively enriched in the sEV fractions. Our results suggest that DNA associated to sEV (including exosomes) is not preferentially enriched in tumor signal and is less abundant than cfDNA.
Keywords: Cancer; Cancer systems biology; Molecular genetics.
© 2024 The Author(s).
Conflict of interest statement
F.M. is coinventor on patents related to cfDNA fragmentation analysis. F.M. has consulted for Roche Dx. D.M.P. is co-founder and CSO of ExBiome BV. Other coauthors have no relevant conflict of interests.
Figures


References
-
- Merker J.D., Oxnard G.R., Compton C., Diehn M., Hurley P., Lazar A.J., Lindeman N., Lockwood C.M., Rai A.J., Schilsky R.L., et al. Circulating Tumor DNA Analysis in Patients With Cancer: American Society of Clinical Oncology and College of American Pathologists Joint Review. J. Clin. Oncol. 2018;36:1631–1641. doi: 10.1200/JCO.2017.76.8671. - DOI - PubMed
LinkOut - more resources
Full Text Sources

