Control of epigenomic landscape and development of fetal male germ cells through L-serine metabolism
- PMID: 39262797
- PMCID: PMC11388182
- DOI: 10.1016/j.isci.2024.110702
Control of epigenomic landscape and development of fetal male germ cells through L-serine metabolism
Abstract
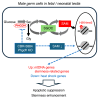
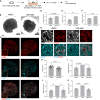
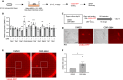
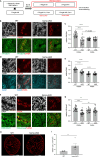
Sex-specific metabolic characteristics emerge in the mouse germ line after reaching the genital ridges around embryonic day 10.5, coinciding with sexual differentiation. However, the impact of such metabolic characteristics on germ cell development remains unclear. In this study, we observed the specific upregulation in male fetal germ cells of D-3-phosphoglycerate dehydrogenase (PHGDH), the primary enzyme in the serine-glycine-one-carbon metabolism, along with an increase in a downstream metabolite, S-adenosylmethionine (SAM), crucial for protein and nucleic acid methylation. Inhibiting PHGDH in fetal testes resulted in reduced SAM levels in germ cells, accompanied by increases in the number of mouse vasa homolog (MVH/VASA)-positive germ cells and the promyelocytic leukemia zinc finger (PLZF)-positive undifferentiated spermatogonia ratio. Furthermore, PHGDH inhibition led to a decrease in the methylation of histone H3 and DNA, resulting in aberrations in gene expression profiles. In summary, our findings underscore the significant role of certain metabolic mechanisms in the development of male germ cells.
Keywords: Cellular physiology; Developmental biology; Epigenetics.
© 2024 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

