Genome of the most noxious weed water hyacinth (Eichhornia crassipes) provides insights into plant invasiveness and its translational potential
- PMID: 39262811
- PMCID: PMC11387899
- DOI: 10.1016/j.isci.2024.110698
Genome of the most noxious weed water hyacinth (Eichhornia crassipes) provides insights into plant invasiveness and its translational potential
Abstract
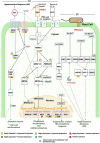
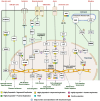
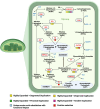
The invasive character of Eichhornia crassipes (water hyacinth) is a major threat to global biodiversity and ecosystems. To investigate the genomic basis of invasiveness, we performed the genome and transcriptome sequencing of E. crassipes and reported the genome of 1.11 Gbp size with 63,299 coding genes and N50 of 1.98 Mb. We confirmed a recent whole genome duplication event in E. crassipes that resulted in high intraspecific collinearity and significant expansion in gene families. Further, the orthologs gene clustering analysis and comparative evolutionary analysis with 14 other aquatic invasive and non-invasive angiosperm species revealed adaptive evolution in genes associated with plant-pathogen interaction, hormone signaling, abiotic stress tolerance, heavy metals sequestration, photosynthesis, and cell wall biosynthesis with highly expanded gene families, which contributes toward invasive characteristics of the water hyacinth. However, these characteristics also make water hyacinth an excellent candidate for biofuel production, phytoremediation, and other translational applications.
Keywords: Genomics; Plant Genetics; Plant biology.
© 2024 Published by Elsevier Inc.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Pyšek P., Richardson D.M. Invasive Species, Environmental Change and Management, and Health. Annu. Rev. Environ. Resour. 2010;35:25–55. doi: 10.1146/annurev-environ-033009-095548. - DOI
-
- Simberloff D., Von Holle B. Positive interactions of nonindigenous species: Invasional meltdown? Biol. Invasions. 1999;1:21–32. doi: 10.1023/A:1010086329619/METRICS. - DOI
-
- Keane R., Crawley M.J. Exotic plant invasions and the enemy release hypothesis. Trends Ecol. Evol. 2002;17:164–170. doi: 10.1016/S0169-5347(02)02499-0. - DOI
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

