Identification and verification of disulfidptosis-related genes in sepsis-induced acute lung injury
- PMID: 39262873
- PMCID: PMC11389619
- DOI: 10.3389/fmed.2024.1430252
Identification and verification of disulfidptosis-related genes in sepsis-induced acute lung injury
Abstract
Background: Sepsis-induced acute lung injury (ALI) is a common and serious complication of sepsis that eventually progresses to life-threatening hypoxemia. Disulfidptosis is a newly discovered type of cell death associated with the pathogenesis of different diseases. This study investigated the potential association between sepsis-induced acute lung injury and disulfidptosis by bioinformatics analysis.
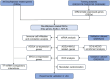
Methods: In order to identify differentially expressed genes (DEGs) linked to sepsis, we screened appropriate data sets from the GEO database and carried out differential analysis. The key genes shared by DEGs and 39 disulfidptosis-related genes were identified: ACSL4 and MYL6 mRNA levels of key genes were detected in different datasets. We then used a series of bioinformatics analysis techniques, such as immune cell infiltration analysis, protein-protein interaction (PPI) network, genetic regulatory network, and receiver operating characteristic (ROC), to investigate the possible relationship between key genes and sepsis. Then, experimental verification was obtained for changes in key genes in sepsis-induced acute lung injury. Finally, to investigate the relationship between genetic variants of MYL6 or ACSL4 and sepsis, Mendelian randomization (MR) analysis was applied.
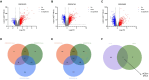
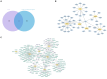
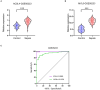
Results: Two key genes were found in this investigation: myosin light chain 6 (MYL6) and Acyl-CoA synthetase long-chain family member 4 (ACSL4). We verified increased mRNA levels of key genes in training datasets. Immune cell infiltration analysis showed that key genes were associated with multiple immune cell levels. Building the PPI network between MYL6 and ACSL4 allowed us to determine that their related genes had distinct biological functions. The co-expression genes of key genes were involved in different genetic regulatory networks. In addition, both the training and validation datasets confirmed the diagnostic capabilities of key genes by using ROC curves. Additionally, both in vivo and in vitro experiments confirmed that the mRNA levels of ACSL4 and MYL6 in sepsis-induced acute lung injury were consistent with the results of bioinformatics analysis. Finally, MR analysis revealed a causal relationship between MYL6 and sepsis.
Conclusion: We have discovered and confirmed that the key genes ACSL4 and MYL6, which are linked to disulfidptosis in sepsis-induced acute lung injury, may be useful in the diagnosis and management of septic acute lung injury.
Keywords: Mendelian randomization; disulfidptosis; immune infiltration; network analysis; sepsis.
Copyright © 2024 Zhang, Wang, Lin, Zhu and Pan.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
LinkOut - more resources
Full Text Sources
Miscellaneous

