Methylation-related differentially expressed genes as potential prognostic biomarkers for cervical cancer
- PMID: 39263148
- PMCID: PMC11387271
- DOI: 10.1016/j.heliyon.2024.e36240
Methylation-related differentially expressed genes as potential prognostic biomarkers for cervical cancer
Abstract
Aim: To discover novel methylation-related differentially expressed genes (MRDEGs) for cervical cancer, with a focus on their potential for early diagnosis and prognostic assessment.
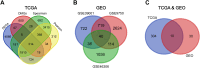
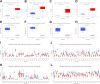
Materials & methods: We integrated data from The Cancer Genome Atlas (TCGA) and Gene Expression Omnibus (GEO) databases. TCGA-MRDEGs were identified by analyzing differentially methylated genes (DMGs) and their correlation with gene expression. We examined GEO datasets GSE39001, GSE9750, and GSE46306 for GEO-MRDEGs. Overlapping MRDEGs were subjected to overall survival (OS) analysis to identify prognostic markers. The expression and methylation levels of these genes were validated in a total of 30 tissue samples, comprising 20 from cervical cancer patients and 10 from normal cervical tissues, using qRT-PCR and MassARRAY EpiTYPER Assay.
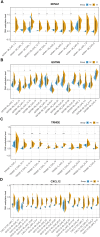
Results: A total of 314 TCGA-MRDEGs and 40 GEO-MRDEGs were identified. Intersection analysis yielded 10 overlapping MRDEGs. Notably, NOVA1, GSTM5, TRHDE, and CXCL12 were found to have reduced expression and increased methylation in cervical cancer, which correlated with poor prognosis. The methylation status and expression levels of these genes were confirmed in tissue specimens.
Conclusion: We identified four MRDEGs as potential prognostic biomarkers for cervical cancer. Their clinical utility is highlighted, but further validation in larger cohorts is required to establish their clinical significance.
Keywords: Biomarker; Cervical cancer; DNA methylation; Differentially expressed genes; Prognosis.
© 2024 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures





References
LinkOut - more resources
Full Text Sources
Other Literature Sources

