Rapid discrimination between wild and cultivated Ophiocordyceps sinensis through comparative analysis of label-free SERS technique and mass spectrometry
- PMID: 39263205
- PMCID: PMC11387260
- DOI: 10.1016/j.crfs.2024.100820
Rapid discrimination between wild and cultivated Ophiocordyceps sinensis through comparative analysis of label-free SERS technique and mass spectrometry
Abstract
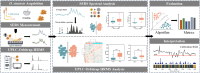
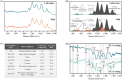
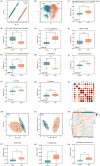
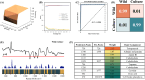
Ophiocordyceps sinensis is a genus of ascomycete fungi that has been widely used as a valuable tonic or medicine. However, due to over-exploitation and the destruction of natural ecosystems, the shortage of wild O. sinensis resources has led to an increase in artificially cultivated O. sinensis. To rapidly and accurately identify the molecular differences between cultivated and wild O. sinensis, this study employs surface-enhanced Raman spectroscopy (SERS) combined with machine learning algorithms to distinguish the two O. sinensis categories. Specifically, we collected SERS spectra for wild and cultivated O. sinensis and validated the metabolic profiles of SERS spectra using Ultra-Performance Liquid Chromatography coupled with Orbitrap High-Resolution Mass Spectrometry (UPLC-Orbitrap-HRMS). Subsequently, we constructed machine learning classifiers to mine potential information from the spectral data, and the spectral feature importance map is determined through an optimized algorithm. The results indicate that the representative characteristic peaks in the SERS spectra are consistent with the metabolites identified through metabolomics analysis, confirming the feasibility of the SERS method. The optimized support vector machine (SVM) model achieved the most accurate and efficient capacity in discriminating between wild and cultivated O. sinensis (accuracy = 98.95%, 5-fold cross-validation = 98.38%, time = 0.89s). The spectral feature importance map revealed subtle compositional differences between wild and cultivated O. sinensis. Taken together, these results are expected to enable the application of SERS in the quality control of O. sinensis raw materials, providing a foundation for the efficient and rapid identification of their quality and origin.
Keywords: Cultivation; Machine learning; Metabolomics; Ophiocordyceps sinensis; Surface-enhanced Raman spectroscopy.
© 2024 The Authors.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Chen H., Tan C., Li H. Discrimination between wild-grown and cultivated Gastrodia elata by near-infrared spectroscopy and chemometrics. Vib. Spectrosc. 2021;113
-
- Chen L., Liu Y., Guo Q., Zheng Q., Zhang W. Metabolomic comparison between wild Ophiocordyceps sinensis and artificial cultured Cordyceps militaris. Biomed. Chromatogr. 2018;32(9) - PubMed
-
- Du C., Zhou J., Liu J. Identification of Chinese medicinal fungus Cordyceps sinensis by depth-profiling mid-infrared photoacoustic spectroscopy. Spectrochim. Acta Mol. Biomol. Spectrosc. 2017;173:489–494. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

