Klebsiella pneumoniae species complex: From wastewater to the environment
- PMID: 39263320
- PMCID: PMC11387367
- DOI: 10.1016/j.onehlt.2024.100880
Klebsiella pneumoniae species complex: From wastewater to the environment
Abstract
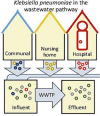
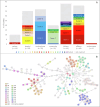
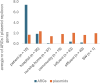
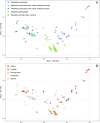
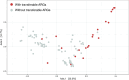
Klebsiella pneumoniae plays a significant role in nosocomial infections and spreading antibiotic resistance, and therefore forms a major threat to public health. In this study, we investigated the role of the wastewater pathway in the spread of pathogenic bacteria and more specifically, in the spread of antibiotic resistant Klebsiella pneumoniae subspecies. Whole-genome sequencing was performed of 185 K. pneumoniae isolates collected from hospital, nursing home, and community wastewater, the receiving wastewater treatment plant (WWTP), and clinical isolates from the investigated hospital. K. pneumoniae isolates from different sources were not genetically related, except for WWTP influent (46.5%) and effluent (62.5%), revealing survival of bacteria from wastewater treatment. The content of antibiotic resistance (ARGs), virulence, and plasmid replicon genes differed between K. pneumoniae subspecies and their origin. While chromosomal bla genes were specific for each K. pneumoniae subspecies, bla genes predicted in plasmid contigs were found in several K. pneumoniae subspecies, implying possible gene transfer between subspecies. Transferable ARGs were most abundant in patients and hospital isolates (70%), but the average number of plasmid replicon genes per isolate was similar across all sources, showing plasmid content being more relevant than plasmid quantity. Most patient (90%) and hospital wastewater (34%) isolates were K. pneumoniae subsp. pneumoniae, and the yersiniabactin cluster genes ybt, fyuA, and irp12 were only found in this subspecies, as were the IncFII(pECLA), IncHI2A, and IncHI2 plasmid replicon genes, suggesting the clinical origin of these type of plasmids.
Keywords: Antibiotic resistance genes; Hospital; Klebsiella pneumoniae; Plasmid replicon genes; Virulence genes; WWTP; Wastewater; Wastewater pathway; Whole genome sequencing.
© 2024 The Authors. Published by Elsevier B.V.
Conflict of interest statement
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
Figures
References
-
- Pendleton J.N., Gorman S.P., Gilmore B.F. Clinical relevance of the ESKAPE pathogens. Expert Rev. Anti-Infect. Ther. 2013;11(3):297–308. - PubMed
-
- Rice L.B. Federal funding for the study of antimicrobial resistance in nosocomial pathogens: no ESKAPE. J. Infect. Dis. 2008;197(8):1079–1081. - PubMed
-
- Wyres K.L., Lam M.M.C., Holt K.E. Population genomics of Klebsiella pneumoniae. Nat. Rev. Microbiol. 2020;18(6):344–359. - PubMed
LinkOut - more resources
Full Text Sources

