Characterization of methicillin resistant Staphylococcus Aureus in municipal wastewater in Finland
- PMID: 39263321
- PMCID: PMC11388770
- DOI: 10.1016/j.onehlt.2024.100881
Characterization of methicillin resistant Staphylococcus Aureus in municipal wastewater in Finland
Abstract
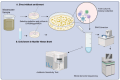
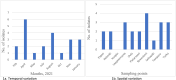
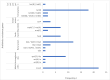
Wastewater-based surveillance (WBS) of multidrug-resistant bacteria could complement clinical data, serving as a population-level early warning tool. This study evaluated WBS as a pandemic preparedness tool, by selectively isolating and culturing methicillin-resistant Staphylococcus aureus (MRSA) with CHROMagar MRSA. Some 24-h composite wastewater samples (n = 80) were collected from ten treatment plants across Finland between February 2021 and January 2022. MRSA prevalence in wastewater samples was 27.5% (n = 22/80), showing seasonal and temporal variations. Phenotypic antimicrobial susceptibility testing (AST) with microdilution showed that over 80% of isolates were drug-resistant to clindamycin, sulfamethoxazole/trimethoprim, tetracycline, fusidic acid, and erythromycin. Four isolates (18.2%) were vancomycin-resistant. WGS revealed that 31.8% (n = 7) of the isolates belonged to the ST8-t008 and ST6-t304 spa types, respectively. In addition, two spa types (t011 and t034) belong to the CC398 complex. The mecA gene was found in all isolates (n = 22) and three tetracycline resistance determinants (tet38, tetK, and tetM) were detected with tet38 being the most abundant (81.8%, n = 18/22). Three isolates harboured the plasmid-mediated sat4 gene that confers resistance to Streptothricin. In addition, resistance determinants to macrolide antibiotics (mph (C)/msr (A) and fosfomycin (fosB) were detected in the seven isolates that belonged to spa type t008. All isolates except one harboured the SCCmec_type_IVa(2B). Six ST8 isolates harboured the LukS/F-PV genes encoding the Panton-Valentine leukocidin (PVL) and were also positive for the Arginine Catabolic Mobile Element (ACME), suggesting they belong to the USA300 clone. The Inc18 plasmid was the most abundant as it was detected in 72.7% (n = 16/22) of the isolates. Other plasmid replicons detected were the rep_trans and repA_N which were detected in 45.4% (n = 10/22) and 40.9% (n = 9/22) of the isolates respectively. Ten isolates harboured at least three plasmid replicons and no plasmid replicons were detected in four isolates (ST6/t304). The cgMLST revealed that some isolates aggregated into two genomically indistinguishable clusters: ST6/t304 belonging to cluster type CT12405 (≤20 allelic differences) and ST8/t008 belonging to cluster type CT1925 (<8 allelic differences). Overall, we found a high genotypic concordance with the national clinical bacterial resistance data. Our study demonstrates the sensitivity of culture-based wastewater surveillance for MRSA using clinical media following pre-enrichment, reliably predicting pathogen occurrence at the population level.
Keywords: Antibiotic resistance gene; Antimicrobial resistance; LukS/F-PV; Staphylococcus aureus; Wastewater based surveillance.
© 2024 The Authors. Published by Elsevier B.V.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. Kirsi-Maarit Lehto and Sami Oikarinen are the stakeholders of GreenSeq Ltd. Finland. All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations or those of the publisher, the editors, or the reviewers.
Figures

References
-
- Petersen A., Larssen K.W., Gran F.W., Enger H., Hæggman S., Mäkitalo B., Haraldsson G., Lindholm L., Vuopio J., Henius A.E., Nielsen J. Increasing incidences and clonal diversity of methicillin-resistant Staphylococcus aureus in the Nordic countries-results from the Nordic MRSA surveillance. Front. Microbiol. 2021 Apr 30;12:668900. - PMC - PubMed
-
- Tiwari A., Krolicka A., Tran T.T., Räisänen K., Ásmundsdóttir Á.M., Wikmark O.G., Lood R., Pitkänen T. Antibiotic resistance monitoring in wastewater in the Nordic countries: a systematic review. Environ. Res. 2024 Apr 1;246:118052. - PubMed
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

