Identification of autophagy-related genes ATG18 subfamily genes in potato (Solanum tuberosum L.) and the role of StATG18a gene in heat stress
- PMID: 39263419
- PMCID: PMC11387889
- DOI: 10.3389/fpls.2024.1439972
Identification of autophagy-related genes ATG18 subfamily genes in potato (Solanum tuberosum L.) and the role of StATG18a gene in heat stress
Abstract
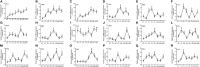
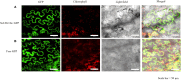
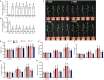
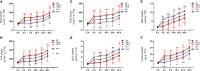
Autophagy is a highly conserved process in eukaryotes that is used to recycle the cellular components from the cytoplasm. It plays a crucial function in responding to both biotic and abiotic stress, as well as in the growth and development of plants. Autophagy-related genes (ATG) and their functions have been identified in numerous crop species. However, their specific tasks in potatoes (Solanum tuberosum L.), are still not well understood. This work is the first to identify and characterize the potato StATG18 subfamily gene at the whole-genome level, resulting in a total of 6 potential StATG18 subfamily genes. We analyzed the phylogenetic relationships, chromosome distribution and gene replication, conserved motifs and gene structure, interspecific collinearity relationship, and cis-regulatory elements of the ATG18 subfamily members using bioinformatics approaches. Furthermore, the quantitative real-time polymerase chain reaction (qRT-PCR) analysis suggested that StATG18 subfamily genes exhibit differential expression in various tissues and organs of potato plants. When exposed to heat stress, their expression pattern was observed in the root, stem, and leaf. Based on a higher expression profile, the StATG18a gene was further analyzed under heat stress in potatoes. The subcellular localization analysis of StATG18a revealed its presence in both the cytoplasm and nucleus. In addition, StATG18a altered the growth indicators, physiological characteristics, and photosynthesis of potato plants under heat stresses. In conclusion, this work offers a thorough assessment of StATG18 subfamily genes and provides essential recommendations for additional functional investigation of autophagy-associated genes in potato plants. Moreover, these results also contribute to our understanding of the potential mechanism and functional validation of the StATG18a gene's persistent tolerance to heat stress in potato plants.
Keywords: StATG18a; autophagy; heat stress; photosynthesis; physiological; potato.
Copyright © 2024 Zhu, Li, Zhang, Duan, Jin, Chen, Chen, Zhou, Wang, Tang, Majeed, Zhang and Si.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Bates L. S., Waldren R. P. A., Teare I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. doi: 10.1007/BF00018060 - DOI
-
- Bouaziz D., Charfeddine M., Jbir R., Saidi M. N., Pirrello J., Charfeddine S., et al. (2015). Identification and functional characterization of ten AP2/ERF genes in potatoes. Plant Cell Tissue Organ Cult. 123, 155–172. doi: 10.1007/s11240-015-0823-2 - DOI
LinkOut - more resources
Full Text Sources

