Mitochondrial genomes of Macropsini (Hemiptera: Cicadellidae: Eurymelinae): Structural features, codon usage patterns, and phylogenetic implications
- PMID: 39263460
- PMCID: PMC11387203
- DOI: 10.1002/ece3.70268
Mitochondrial genomes of Macropsini (Hemiptera: Cicadellidae: Eurymelinae): Structural features, codon usage patterns, and phylogenetic implications
Abstract
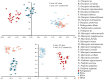
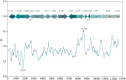
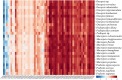
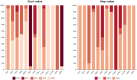
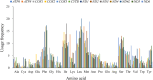
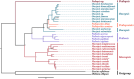
Macropsini is a tribe of Eurymelinae in the family Cicadellidae that is widely distributed worldwide. Still, its taxonomic status has been unstable, and the classification of certain clades at the genus level has been controversial. The aim of this study is to address the patterns and processes that explain the structure and the evolution of the mitogenomes of Macropsini, while contributing to the resolution of systematic issues involving five of their genera. To this task, the mitogenomes of 26 species of the tribe were sequenced and characterized, and their phylogenetic relationships were reconstructed. The results revealed that the nucleotide composition of mitochondrial genes in these 26 species was significantly skewed toward A and T. Codons ending with T or A in relative synonymous codon usage were significantly more prevalent than those ending with C or G. The parity plot, neutrality plot, and correspondence analysis revealed that mutation and selective pressure affect codon usage patterns. In the phylogenetic relationships of the Macropsini, the monophyly of Pedionis and Macropsis was well-supported. Meanwhile, Oncopsis revealed paraphyletic regarding Pediopsoides. In conclusion, this research not only contributes the valuable data to the understanding of the mitogenome of the Macropsini but also provides a reference for future investigations on codon usage patterns, potential adaptive evolution, and the phylogeny of the mitogenome within the subfamily Eurymelinae.
Keywords: Macropsini; codon usage bias; mutation pressure; natural selection; phylogeny.
© 2024 The Author(s). Ecology and Evolution published by John Wiley & Sons Ltd.
Conflict of interest statement
All authors declare no conflict of interest.
Figures




Similar articles
-
Comparative analysis of codon usage patterns and phylogenetic implications of five mitochondrial genomes of the genus Japanagallia Ishihara, 1955 (Hemiptera, Cicadellidae, Megophthalminae).PeerJ. 2023 Sep 25;11:e16058. doi: 10.7717/peerj.16058. eCollection 2023. PeerJ. 2023. PMID: 37780390 Free PMC article.
-
Description of mitochondrial genomes and phylogenetic analysis of Megophthalminae (Hemiptera: Cicadellidae).J Insect Sci. 2024 Nov 1;24(6):9. doi: 10.1093/jisesa/ieae109. J Insect Sci. 2024. PMID: 39657582 Free PMC article.
-
Structural Characteristics and Phylogenetic Analysis of the Mitochondrial Genomes of Four Krisna Species (Hemiptera: Cicadellidae: Iassinae).Genes (Basel). 2023 May 28;14(6):1175. doi: 10.3390/genes14061175. Genes (Basel). 2023. PMID: 37372355 Free PMC article.
-
Structural features of the mitogenome of the leafhopper genus Cladolidia (Hemiptera: Cicadellidae: Coelidiinae) and phylogenetic implications in Cicadellidae.Ecol Evol. 2021 Aug 5;11(18):12554-12566. doi: 10.1002/ece3.8001. eCollection 2021 Sep. Ecol Evol. 2021. PMID: 34594520 Free PMC article.
-
The mitochondrial genome sequences of eleven leafhopper species of Batracomorphus (Hemiptera: Cicadellidae: Iassinae) reveal new gene rearrangements and phylogenetic implications.PeerJ. 2024 Oct 22;12:e18352. doi: 10.7717/peerj.18352. eCollection 2024. PeerJ. 2024. PMID: 39465150 Free PMC article.
Cited by
-
Comparative Analysis of Codon Usage Bias in Transcriptomes of Eight Species of Formicidae.Genes (Basel). 2025 Jun 27;16(7):749. doi: 10.3390/genes16070749. Genes (Basel). 2025. PMID: 40725406 Free PMC article.
References
-
- Ballard, J. W. , & Whitlock, M. C. (2004). The incomplete natural history of mitochondria. Molecular Ecology, 13(4), 729–744. - PubMed
-
- Beirne, B. P. (1954). The Prunus‐ and Rubus‐feeding species of Macropsis (Homoptera: Cicadellidae). The Canadian Entomologist, 86(2), 86–90.
-
- Bernt, M. , Donath, A. , Jühling, F. , Externbrink, F. , Florentz, C. , Fritzsch, G. , Pütz, J. , Middendorf, M. , & Stadler, P. F. (2012). MITOS: Improved de novo metazoan mitochondrial genome annotation. Molecular Phylogenetics and Evolution, 69(2), 313–319. - PubMed
-
- Cameron, S. L. (2013). Insect mitochondrial genomics: Implications for evolution and phylogeny. Annual Review of Entomology, 59, 95–117. - PubMed
LinkOut - more resources
Full Text Sources

