Rational Design of Lipid Nanoparticles for Enhanced mRNA Vaccine Delivery via Machine Learning
- PMID: 39264000
- PMCID: PMC11855220
- DOI: 10.1002/smll.202405618
Rational Design of Lipid Nanoparticles for Enhanced mRNA Vaccine Delivery via Machine Learning
Abstract
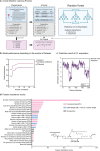
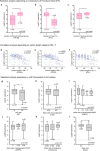
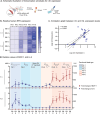
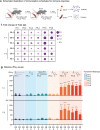
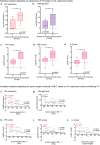
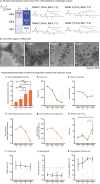
Since the coronavirus pandemic, mRNA vaccines have revolutionized the field of vaccinology. Lipid nanoparticles (LNPs) are proposed to enhance mRNA delivery efficiency; however, their design is suboptimal. Here, a rational method for designing LNPs is explored, focusing on the ionizable lipid composition and structural optimization using machine learning (ML) techniques. A total of 213 LNPs are analyzed using random forest regression models trained with 314 features to predict the mRNA expression efficiency. The models, which predict mRNA expression levels post-administration of intradermal injection in mice, identify phenol as the dominant substructure affecting mRNA encapsulation and expression. The specific phospholipids used as components of the LNPs, as well as the N/P ratio and mass ratio, are found to affect the efficacy of mRNA delivery. Structural analysis highlights the impact of the carbon chain length on the encapsulation efficiency and LNP stability. This integrated approach offers a framework for designing advanced LNPs and has the potential to unlock the full potential of mRNA therapeutics.
Keywords: lipid nanoparticles (LNPs); mRNA expression efficiency; mRNA vaccines; machine learning (ML) techniques; structural optimization.
© 2024 The Author(s). Small published by Wiley‐VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Xu Y., Golubovic A., Xu S., Pan A., Li B., J. Mater. Chem. B 2023, 11, 6527. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

