Antibody sequence determinants of viral antigen specificity
- PMID: 39264172
- PMCID: PMC11481873
- DOI: 10.1128/mbio.01560-24
Antibody sequence determinants of viral antigen specificity
Abstract
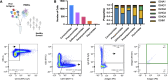
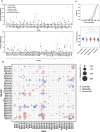
Throughout life, humans experience repeated exposure to viral antigens through infection and vaccination, resulting in the generation of diverse, antigen-specific antibody repertoires. A paramount feature of antibodies that enables their critical contributions in counteracting recurrent and novel pathogens, and consequently fostering their utility as valuable targets for therapeutic and vaccine development, is the exquisite specificity displayed against their target antigens. Yet, there is still limited understanding of the determinants of antibody-antigen specificity, particularly as a function of antibody sequence. In recent years, experimental characterization of antibody repertoires has led to novel insights into fundamental properties of antibody sequences but has been largely decoupled from at-scale antigen specificity analysis. Here, using the LIBRA-seq technology, we generated a large data set mapping antibody sequence to antigen specificity for thousands of B cells, by screening the repertoires of a set of healthy individuals against 20 viral antigens representing diverse pathogens of biomedical significance. Analysis uncovered virus-specific patterns in variable gene usage, gene pairing, somatic hypermutation, as well as the presence of convergent antiviral signatures across multiple individuals, including the presence of public antibody clonotypes. Notably, our results showed that, for B-cell receptors originating from different individuals but leveraging an identical combination of heavy and light chain variable genes, there is a specific CDRH3 identity threshold above which B cells appear to exclusively share the same antigen specificity. This finding provides a quantifiable measure of the relationship between antibody sequence and antigen specificity and further defines experimentally grounded criteria for defining public antibody clonality.IMPORTANCEThe B-cell compartment of the humoral immune system plays a critical role in the generation of antibodies upon new and repeated pathogen exposure. This study provides an unprecedented level of detail on the molecular characteristics of antibody repertoires that are specific to each of the different target pathogens studied here and provides empirical evidence in support of a 70% CDRH3 amino acid identity threshold in pairs of B cells encoded by identical IGHV:IGL(K)V genes, as a means of defining public clonality and therefore predicting B-cell antigen specificity in different individuals. This is of exceptional importance when leveraging public clonality as a method to annotate B-cell receptor data otherwise lacking antigen specificity information. Understanding the fundamental rules of antibody-antigen interactions can lead to transformative new approaches for the development of antibody therapeutics and vaccines against current and emerging viruses.
Keywords: B-cell repertoire; B-cell responses; antibody repertoire; antigen specificity; public clonotype; virus.
Conflict of interest statement
A.A.A.-S. and I.S.G. are listed as inventors on patents filed describing the antibodies discovered here. I.S.G. is listed as an inventor on patent applications for the LIBRA-seq technology. I.S.G. is a co-founder of AbSeek Bio. I.S.G. has served as a consultant for Sanofi. The Georgiev laboratory at VUMC has received unrelated funding from Merck and Takeda Pharmaceuticals.
Figures


References
-
- Shrock EL, Timms RT, Kula T, Mena EL, West AP, Guo R, Lee IH, Cohen AA, McKay LGA, Bi C, Leng Y, Fujimura E, Horns F, Li M, Wesemann DR, Griffiths A, Gewurz BE, Bjorkman PJ, Elledge SJ. 2023. Germline-encoded amino acid-binding motifs drive immunodominant public antibody responses. Science 380:eadc9498. doi: 10.1126/science.adc9498 - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
- R01 AI152693/AI/NIAID NIH HHS/United States
- VR54735/Vanderbilt Institute for Clinical and Translational Research (VICTR)
- T15 LM007450/LM/NLM NIH HHS/United States
- P30 DK058404/DK/NIDDK NIH HHS/United States
- T15LM007450-19/HHS | NIH | National Institute of Environmental Health Sciences (NIEHS)
- ZIAAI005003/HHS | National Institutes of Health (NIH)
- P30 EY008126/EY/NEI NIH HHS/United States
- ZIA AI005003/ImNIH/Intramural NIH HHS/United States
- R01AI175245/HHS | National Institutes of Health (NIH)
- UL1 RR024975/RR/NCRR NIH HHS/United States
- UL1 TR002243/TR/NCATS NIH HHS/United States
- F-0003-19620604/Welch Foundation (The Welch Foundation)
- 5T32AI112541-07/HHS | National Institutes of Health (NIH)
- MF-2107-01851/G. Harold and Leila Y. Mathers Foundation (Mathers Foundation)
- R01 AI175245/AI/NIAID NIH HHS/United States
- T32 AI112541/AI/NIAID NIH HHS/United States
- G20 RR030956/RR/NCRR NIH HHS/United States
- P30 CA068485/CA/NCI NIH HHS/United States
- P30 AI110527/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

