Midnolin, a Genetic Risk Factor for Parkinson's Disease, Promotes Neurite Outgrowth Accompanied by Early Growth Response 1 Activation in PC12 Cells
- PMID: 39264361
- PMCID: PMC11529416
- DOI: 10.1080/10985549.2024.2399358
Midnolin, a Genetic Risk Factor for Parkinson's Disease, Promotes Neurite Outgrowth Accompanied by Early Growth Response 1 Activation in PC12 Cells
Abstract
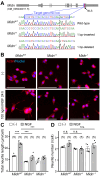
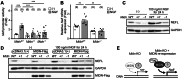
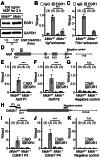
Parkinson's disease (PD) is an age-related progressive neurodegenerative disease. Previously, we identified midnolin (MIDN) as a genetic risk factor for PD. Although MIDN copy number loss increases the risk of PD, the molecular function of MIDN remains unclear. To investigate the role of MIDN in PD, we established monoclonal Midn knockout (KO) PC12 cell models. Midn KO inhibited neurite outgrowth and neurofilament light chain (Nefl) gene expression. Although MIDN is mainly localized in the nucleus, it does not encode DNA-binding domains. We therefore hypothesized that MIDN might bind to certain transcription factors and regulate gene expression. Of the candidate transcription factors, we focused on early growth response 1 (EGR1) because it is required for neurite outgrowth and its target genes are downregulated by Midn KO. An interaction between MIDN and EGR1 was confirmed by immunoprecipitation. Surprisingly, although EGR1 protein levels were significantly increased in Midn KO cells, the binding of EGR1 to the Nefl promoter and resulting transcriptional activity were downregulated as measured by luciferase assay and chromatin immunoprecipitation quantitative real-time polymerase chain reaction. Overall, we identified the MIDN-dependent regulation of EGR1 function. This mechanism may be an underlying reason for the neurite outgrowth defects of Midn KO PC12 cells.
Keywords: PC12 cells; Parkinson’s disease (PD); early growth response 1 (EGR1); neurite outgrowth; neurofilament light chain (NEFL).
Conflict of interest statement
Yoshishiro Koyama is an employee of ThermoFisher Scientific KK (Tokyo, Japan) and contributed to the measurement of neurite outgrowth. Apart from this, the authors report that there are no competing interests to declare.
Figures



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
