Pharmacogenomics of clinical response to Natalizumab in multiple sclerosis: a genome-wide multi-centric association study
- PMID: 39264442
- PMCID: PMC11561017
- DOI: 10.1007/s00415-024-12608-6
Pharmacogenomics of clinical response to Natalizumab in multiple sclerosis: a genome-wide multi-centric association study
Erratum in
-
Correction: Pharmacogenomics of clinical response to Natalizumab in multiple sclerosis: a genome-wide multi-centric association study.J Neurol. 2025 May 27;272(6):423. doi: 10.1007/s00415-025-13108-x. J Neurol. 2025. PMID: 40423827 Free PMC article. No abstract available.
Abstract
Background: Inter-individual differences in treatment response are marked in multiple sclerosis (MS). This is true for Natalizumab (NTZ), to which a subset of patients displays sub-optimal treatment response. We conducted a multi-centric genome-wide association study (GWAS), with additional pathway and network analysis to identify genetic predictors of response to NTZ.
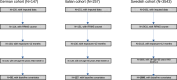
Methods: MS patients from three different centers were included. Response to NTZ was dichotomized, nominating responders (R) relapse-free patients and non-responders (NR) all the others, over a follow-up of 4 years. Association analysis on ~ 4.7 M imputed autosomal common single-nucleotide polymorphisms (SNPs) was performed fitting logistic regression models, adjusted for baseline covariates, followed by meta-analysis at SNP and gene level. Finally, these signals were projected onto STRING interactome, to elicit modules and hub genes linked to response.
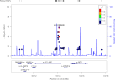
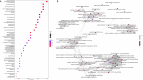
Results: Overall, 1834 patients were included: 119 from Italy (R = 94, NR = 25), 81 from Germany (R = 61, NR = 20), and 1634 from Sweden (R = 1349, NR = 285). The top-associated variant was rs11132400T (p = 1.33 × 10-6, OR = 0.58), affecting expression of several genes in the locus, like KLKB1. The interactome analysis implicated a module of 135 genes, with over-representation of terms like canonical WNT signaling pathway (padjust = 7.08 × 10-6). Response-associated genes like GRB2 and LRP6, already implicated in MS pathogenesis, were topologically prioritized within the module.
Conclusion: This GWAS, the largest pharmacogenomic study of response to NTZ, suggested MS-implicated genes and Wnt/β-catenin signaling pathway, an essential component for blood-brain barrier formation and maintenance, to be related to treatment response.
Keywords: GRB2; LRP6; Multiple sclerosis; Natalizumab; Pharmacogenomics.
© 2024. The Author(s).
Conflict of interest statement
Figures


References
-
- Hauser SL, Oksenberg JR (2006) The neurobiology of multiple sclerosis: genes, inflammation, and neurodegeneration. Neuron 52:61–76. 10.1016/j.neuron.2006.09.011 - PubMed
-
- Grossman I, Knappertz V, Laifenfeld D et al (2017) Pharmacogenomics strategies to optimize treatments for multiple sclerosis: insights from clinical research. Prog Neurobiol 152:114–130. 10.1016/j.pneurobio.2016.02.001 - PubMed
-
- Polman CH, O’Connor PW, Havrdova E et al (2006) A randomized, placebo-controlled trial of natalizumab for relapsing multiple sclerosis. N Engl J Med 354:899–910. 10.1056/NEJMoa044397 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

