Characterization of SMA type II skeletal muscle from treated patients shows OXPHOS deficiency and denervation
- PMID: 39264856
- PMCID: PMC11530132
- DOI: 10.1172/jci.insight.180992
Characterization of SMA type II skeletal muscle from treated patients shows OXPHOS deficiency and denervation
Abstract
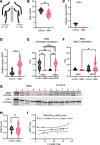
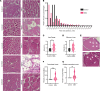
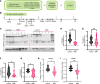
Spinal muscular atrophy (SMA) is a recessive developmental disorder caused by the genetic loss or mutation of the gene SMN1 (survival of motor neuron 1). SMA is characterized by neuromuscular symptoms and muscle weakness. Several years ago, SMA treatment underwent a radical transformation, with the approval of 3 different SMN-dependent disease-modifying therapies. This includes 2 SMN2 splicing therapies - risdiplam and nusinersen. One main challenge for type II SMA patients treated with these drugs is ongoing muscle fatigue, limited mobility, and other skeletal problems. To date, few molecular studies have been conducted on SMA patient-derived tissues after treatment, limiting our understanding of what targets remain unchanged after the spinal cord-targeted therapies are applied. Therefore, we collected paravertebral muscle from 8 type II patients undergoing spinal surgery for scoliosis and 7 controls. We used RNA-seq to characterize their transcriptional profiles and correlate these molecular changes with muscle histology. Despite the limited cohort size and heterogeneity, we observed a consistent loss of oxidative phosphorylation (OXPHOS) machinery of the mitochondria, a decrease in mitochondrial DNA copy number, and a correlation between signals of cellular stress, denervation, and increased fibrosis. This work provides new putative targets for combination therapies for type II SMA.
Keywords: Bioinformatics; Genetics; Muscle biology; Neuromuscular disease; Skeletal muscle.
Conflict of interest statement
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

