Drivers of diversification in fungal pathogen populations
- PMID: 39264909
- PMCID: PMC11392411
- DOI: 10.1371/journal.ppat.1012430
Drivers of diversification in fungal pathogen populations
Abstract
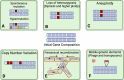
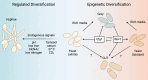
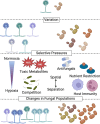
To manage and treat chronic fungal diseases effectively, we require an improved understanding of their complexity. There is an increasing appreciation that chronic infection populations are often heterogeneous due to diversification and drift, even within a single microbial species. Genetically diverse populations can contribute to persistence and resistance to treatment by maintaining cells with different phenotypes capable of thriving in these dynamic environments. In chronic infections, fungal pathogens undergo prolonged challenges that can drive trait selection to convergent adapted states through restricted access to critical nutrients, assault by immune effectors, competition with other species, and antifungal drugs. This review first highlights the various genetic and epigenetic mechanisms that promote diversity in pathogenic fungal populations and provide an additional barrier to assessing the actual heterogeneity of fungal infections. We then review existing studies of evolution and genetic heterogeneity in fungal populations from lung infections associated with the genetic disease cystic fibrosis. We conclude with a discussion of open research questions that, once answered, may aid in diagnosing and treating chronic fungal infections.
Copyright: © 2024 Murante, Hogan. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
Similar articles
-
Facilitators of adaptation and antifungal resistance mechanisms in clinically relevant fungi.Fungal Genet Biol. 2019 Nov;132:103254. doi: 10.1016/j.fgb.2019.103254. Epub 2019 Jul 19. Fungal Genet Biol. 2019. PMID: 31326470 Review.
-
Developing collaborative works for faster progress on fungal respiratory infections in cystic fibrosis.Med Mycol. 2018 Apr 1;56(suppl_1):42-59. doi: 10.1093/mmy/myx106. Med Mycol. 2018. PMID: 29538733 Review.
-
Aerially transmitted human fungal pathogens: what can we learn from metagenomics and comparative genomics?Rev Iberoam Micol. 2014 Jan-Mar;31(1):54-61. doi: 10.1016/j.riam.2013.10.006. Epub 2013 Nov 26. Rev Iberoam Micol. 2014. PMID: 24286763 Review.
-
Seasons of change: Mechanisms of genome evolution in human fungal pathogens.Infect Genet Evol. 2019 Jun;70:165-174. doi: 10.1016/j.meegid.2019.02.031. Epub 2019 Mar 1. Infect Genet Evol. 2019. PMID: 30826447 Review.
-
Immunometabolism in fungal infections: the need to eat to compete.Curr Opin Microbiol. 2020 Dec;58:32-40. doi: 10.1016/j.mib.2020.07.001. Epub 2020 Aug 9. Curr Opin Microbiol. 2020. PMID: 32781324 Review.
Cited by
-
A general and biomedical perspective of viral quasispecies.RNA. 2025 Feb 19;31(3):429-443. doi: 10.1261/rna.080280.124. RNA. 2025. PMID: 39689947 Free PMC article. Review.
-
Genome restructuring and lineage diversification of Cryptococcus neoformans during chronic infection of human hosts.medRxiv [Preprint]. 2025 Feb 21:2025.02.17.25320472. doi: 10.1101/2025.02.17.25320472. medRxiv. 2025. Update in: Cell Rep. 2025 Aug 5;44(8):116103. doi: 10.1016/j.celrep.2025.116103. PMID: 40034768 Free PMC article. Updated. Preprint.
-
Genome restructuring and lineage diversification of Cryptococcus neoformans during chronic infection of human hosts.Cell Rep. 2025 Aug 5;44(8):116103. doi: 10.1016/j.celrep.2025.116103. Online ahead of print. Cell Rep. 2025. PMID: 40768337 Free PMC article.
References
-
- Dennis EK, Garneau-Tsodikova S. Synergistic combinations of azoles and antihistamines against Candida species in vitro. Med Mycol. 2019;57:874–884. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

