Divergent effects of acute and chronic PPT1 inhibition in melanoma
- PMID: 39265628
- PMCID: PMC11760279
- DOI: 10.1080/15548627.2024.2403152
Divergent effects of acute and chronic PPT1 inhibition in melanoma
Abstract
Macroautophagy/autophagy-lysosome function promotes growth and survival of cancer cells, making them attractive targets for cancer therapy. One intriguing lysosomal target is PPT1 (palmitoyl-protein thioesterase 1). PPT1 inhibitors derived from chloroquine block autophagy, have significant antitumor activity in preclinical models and are being developed for clinical trials. However, the role of PPT1 in tumorigenesis remains poorly understood. Here we report that in melanoma cells, acute siRNA or pharmacological PPT1 inhibition led to increased ferroptosis sensitivity and significant loss of viability, whereas chronic PPT1 knockout using CRISPR-Cas9 produced blunted ferroptosis that led to sustained viability and growth. Each mode of PPT1 inhibition produced lysosome-autophagy inhibition but distinct proteomic changes, demonstrating the complexity of cellular adaptation mechanisms. To determine whether total genetic loss of Ppt1 would affect tumorigenesis in vivo, we developed a Ppt1 conditional knockout mouse model. We then crossed it into the BrafCA, PtenloxP, Tyr:CreERT2 melanoma mouse model to investigate the impact of Ppt1 loss on tumorigenesis. Loss of Ppt1 had no impact on melanoma histology, time to tumor initiation, or survival of tumor-bearing mice. These results suggest that chemical PPT1 inhibitors produce different adaptations than genetic PPT1 inhibition, and additional studies are warranted to fully understand the mechanism of chloroquine derivatives that target PPT1 in cancer.Abbreviations: 4-HT: 4-hydroxytamoxifen; BRAF: B-Raf proto-oncogene, serine/threonine kinase; cKO: conditional knockout; CRISPR-Cas9: clustered regularly interspaced short palindromic repeats-CRISPR-associated protein 9; DC661: A specific PPT1 inhibitor; DMSO: dimethyl sulfoxide; Dox; doxycycline hyclate; Easi-CRISPR: efficient additions with ssDNA inserts-CRISPR; GNS561/ezurpimtrostat: A PPT1 inhibitor; Hug: human guide; iCas: inducible CRISPR-Cas9; KO: knockout; LC-MS/MS: Liquid chromatography-tandem mass spectrometry; LDLR: low density lipoprotein receptor; NFE2L2/NRF2: NFE2 like bZIP transcription factor 2; NT: non-target; PTEN: phosphatase and tensin homolog; PPT1: palmitoyl-protein thioesterase 1; RSL3: RAS-selective lethal small molecule 3; SCRIB/SCRB1: scribble planar cell polarity protein; Tyr:CreERT2: tyrosinase-driven Cre recombinase fused with the tamoxifen-inducible mutant ligand binding domain of the human estrogen receptor; UGCG: UDP-glucose ceramide glucosyltransferase; WT: wild-type.
Keywords: Autophagy; ferroptosis; lysosome; mouse model; palmitoyl protein thioesterase 1.
Conflict of interest statement
RKA is an inventor on patents related to dimeric chloroquines. The patents are licensed to Pinpoint Therapeutics, and RKA is a scientific founder. RKA is a consultant for Deciphera, Tasca Therapeutics, and gets research funding from Novartis, Bristol-Myers Squibb, Deciphera, Springworks, Merck. The other authors declare no conflicts of interest.
Figures
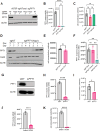
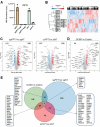
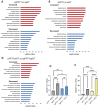

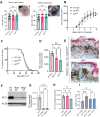
References
-
- Yang A, Herter-Sprie G, Zhang H, et al. Autophagy sustains pancreatic cancer growth through both cell-autonomous and nonautonomous mechanisms. Cancer Discov. 2018. Mar;8(3):276–287. doi: 10.1158/2159-8290.CD-17-0952 - DOI - PMC - PubMed
-
- Xie X, Koh JY, Price S, et al. Atg7 overcomes senescence and promotes growth of BrafV600E-Driven melanoma. Cancer Discov. 2015. Apr;5(4):410–423. doi: 10.1158/2159-8290.CD-14-1473 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous
