A metabolic crosstalk between liposarcoma and muscle sustains tumor growth
- PMID: 39266552
- PMCID: PMC11393074
- DOI: 10.1038/s41467-024-51827-3
A metabolic crosstalk between liposarcoma and muscle sustains tumor growth
Abstract
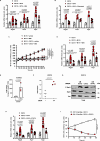
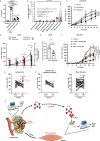
Dedifferentiated and Well-differentiated liposarcoma are characterized by a systematic amplification of the Murine Double Minute 2 (MDM2) oncogene. We demonstrate that p53-independent metabolic functions of chromatin-bound MDM2 are exacerbated in liposarcoma and mediate an addiction to serine metabolism to sustain tumor growth. However, the origin of exogenous serine remains unclear. Here, we show that elevated serine levels in mice harboring liposarcoma-patient derived xenograft, released by distant muscle is essential for liposarcoma cell survival. Repressing interleukine-6 expression, or treating liposarcoma cells with Food and Drugs Administration (FDA) approved anti-interleukine-6 monoclonal antibody, decreases de novo serine synthesis in muscle, impairs proliferation, and increases cell death in vitro and in vivo. This work reveals a metabolic crosstalk between muscle and liposarcoma tumor and identifies anti-interleukine-6 as a plausible treatment for liposarcoma patients.
© 2024. The Author(s).
Conflict of interest statement
L.K.L., N.F., and G.M. are inventors on patent application no. 21 306099.9 submitted by ICM that covers “methods for the treatment of cancer”. All other authors declare that they have no competing interests.
Figures



References
-
- Verweij, J. & Baker, L. H. Future treatment of soft tissue sarcomas will be driven by histological subtype and molecular aberrations. Eur. J. Cancer Oxf. Engl. 199046, 863–868 (2010). - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

