Transiently formed nucleus-to-cilium microtubule arrays mediate senescence initiation in a KIFC3-dependent manner
- PMID: 39266565
- PMCID: PMC11393428
- DOI: 10.1038/s41467-024-52363-w
Transiently formed nucleus-to-cilium microtubule arrays mediate senescence initiation in a KIFC3-dependent manner
Erratum in
-
Author Correction: Transiently formed nucleus-to-cilium microtubule arrays mediate senescence initiation in a KIFC3-dependent manner.Nat Commun. 2024 Oct 29;15(1):9331. doi: 10.1038/s41467-024-53795-0. Nat Commun. 2024. PMID: 39472588 Free PMC article. No abstract available.
Abstract
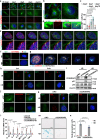
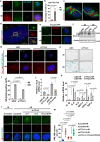
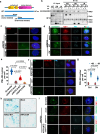
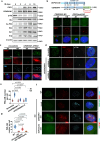
Despite the importance of cellular senescence in human health, how damaged cells undergo senescence remains elusive. We have previously shown that promyelocytic leukemia nuclear body (PML-NBs) translocation of the ciliary FBF1 is essential for senescence induction in stressed cells. Here we discover that an early cellular event occurring in stressed cells is the transient assembly of stress-induced nucleus-to-cilium microtubule arrays (sinc-MTs). The sinc-MTs are distinguished by unusual polyglutamylation and unique polarity, with minus-ends nucleating near the nuclear envelope and plus-ends near the ciliary base. KIFC3, a minus-end-directed kinesin, is recruited to plus-ends of sinc-MTs and interacts with the centrosomal protein CENEXIN1. In damaged cells, CENEXIN1 co-translocates with FBF1 to PML-NBs. Deficiency of KIFC3 abolishes PML-NB translocation of FBF1 and CENEXIN1, as well as senescence initiation in damaged cells. Our study reveals that KIFC3-mediated nuclear transport of FBF1 along polyglutamylated sinc-MTs is a prerequisite for senescence induction in mammalian cells.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
- R01AG076469/Foundation for the National Institutes of Health (Foundation for the National Institutes of Health, Inc.)
- R01 DK099160/DK/NIDDK NIH HHS/United States
- P30 DK090728/DK/NIDDK NIH HHS/United States
- P30 DK090868/DK/NIDDK NIH HHS/United States
- R01 DK139607/DK/NIDDK NIH HHS/United States
- R01DK099160/Foundation for the National Institutes of Health (Foundation for the National Institutes of Health, Inc.)
- Donald E. Wesson Research Fellowship Award/ASN Foundation for Kidney Research (ASN Foundation)
- P30 CA015083/CA/NCI NIH HHS/United States
- Donald E. Wesson Research Fellowship Award/American Society of Nephrology (ASN)
- R01 DK090038/DK/NIDDK NIH HHS/United States
- R01 AG076469/AG/NIA NIH HHS/United States
- W81XWH2010214/U.S. Department of Defense (United States Department of Defense)
- R01DK090038/Foundation for the National Institutes of Health (Foundation for the National Institutes of Health, Inc.)
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

