Dual blockade of IL-10 and PD-1 leads to control of SIV viral rebound following analytical treatment interruption
- PMID: 39266691
- PMCID: PMC11436369
- DOI: 10.1038/s41590-024-01952-4
Dual blockade of IL-10 and PD-1 leads to control of SIV viral rebound following analytical treatment interruption
Erratum in
-
Author Correction: Dual blockade of IL-10 and PD-1 leads to control of SIV viral rebound following analytical treatment interruption.Nat Immunol. 2025 Mar;26(3):524. doi: 10.1038/s41590-025-02079-w. Nat Immunol. 2025. PMID: 39821151 Free PMC article. No abstract available.
Abstract
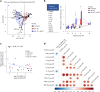
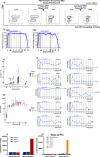
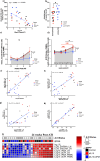
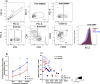
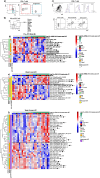
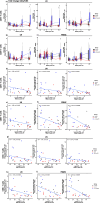
Human immunodeficiency virus (HIV) persistence during antiretroviral therapy (ART) is associated with heightened plasma interleukin-10 (IL-10) levels and PD-1 expression. We hypothesized that IL-10 and PD-1 blockade would lead to control of viral rebound following analytical treatment interruption (ATI). Twenty-eight ART-treated, simian immunodeficiency virus (SIV)mac239-infected rhesus macaques (RMs) were treated with anti-IL-10, anti-IL-10 plus anti-PD-1 (combo) or vehicle. ART was interrupted 12 weeks after introduction of immunotherapy. Durable control of viral rebound was observed in nine out of ten combo-treated RMs for >24 weeks post-ATI. Induction of inflammatory cytokines, proliferation of effector CD8+ T cells in lymph nodes and reduced expression of BCL-2 in CD4+ T cells pre-ATI predicted control of viral rebound. Twenty-four weeks post-ATI, lower viral load was associated with higher frequencies of memory T cells expressing TCF-1 and of SIV-specific CD4+ and CD8+ T cells in blood and lymph nodes of combo-treated RMs. These results map a path to achieve long-lasting control of HIV and/or SIV following discontinuation of ART.
© 2024. The Author(s).
Conflict of interest statement
G.R., Z.S., E.R., G.A., G.W., L.M., D.M.G., B.J.H. and D.J.H. are employed by and/or have financial interests in Merck & Co., Inc. A.-N.P. is a paid consultant, shareholder and board member of RPM Bioinfo Solutions. The other authors declare no competing interests.
Figures










References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

