Inhibition of nucleo-cytoplasmic proteasome translocation by the aromatic amino acids or silencing Sestrin3-their sensing mediator-is tumor suppressive
- PMID: 39266717
- PMCID: PMC11445514
- DOI: 10.1038/s41418-024-01370-x
Inhibition of nucleo-cytoplasmic proteasome translocation by the aromatic amino acids or silencing Sestrin3-their sensing mediator-is tumor suppressive
Abstract
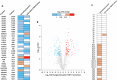
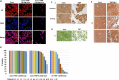
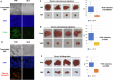
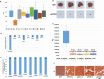
The proteasome, the catalytic arm of the ubiquitin system, is regulated via its dynamic compartmentation between the nucleus and the cytoplasm, among other mechanisms. Under amino acid shortage, the proteolytic complex is translocated to the cytoplasm, where it stimulates proteolysis to supplement recycled amino acids for essential protein synthesis. This response is mediated via the mTOR pathway and the lack of the three aromatic amino acids Tyr, Trp, and Phe (YWF). mTOR activation by supplementation of the triad inhibits proteasome translocation, leading to cell death. We now show that tumoral inherent stress conditions result in translocation of the proteasome from the nucleus to the cytosol. We further show that the modulation of the signaling cascade governed by YWF is applicable also to non-starved cells by using higher concentration of the triad to achieve a surplus relative to all other amino acids. Based on these two phenomena, we found that the modulation of stress signals via the administration of YWF leads to nuclear proteasome sequestration and inhibition of growth of xenograft, spontaneous, and metastatic mouse tumor models. In correlation with the observed effect of YWF on tumors, we found - using transcriptomic and proteomic analyses - that the triad affects various cellular processes related to cell proliferation, migration, and death. In addition, Sestrin3-a mediator of YWF sensing upstream of mTOR-is essential for proteasome translocation, and therefore plays a pro-tumorigenic role, positioning it as a potential oncogene. This newly identified approach for hijacking the cellular "satiety center" carries therefore potential therapeutic implications for cancer.
© 2024. The Author(s).
Conflict of interest statement
IL and AC are co-founders of Tripod Therapeutics Ltd., which develops YWF-based therapies.
Figures



References
-
- Zheng N, Shabek N. Ubiquitin ligases: structure, function, and regulation. 2017. 10.1146/annurev-biochem - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

