Safety and antiviral effect of a triple combination of HIV-1 broadly neutralizing antibodies: a phase 1/2a trial
- PMID: 39266747
- PMCID: PMC11645281
- DOI: 10.1038/s41591-024-03247-5
Safety and antiviral effect of a triple combination of HIV-1 broadly neutralizing antibodies: a phase 1/2a trial
Abstract
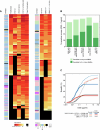
Human immunodeficiency virus type 1 (HIV-1)-specific broadly neutralizing monoclonal antibodies (bNAbs) have to date shown transient viral suppression when administered as monotherapy or as a cocktail of two antibodies1-4. A combination of three bNAbs provides improved neutralization coverage of global viruses, which may more potently suppress viral escape and rebound5-7. Here we performed an open-label, two-part study evaluating a single intravenous dose of HIV-1 bNAbs, PGT121, PGDM1400 and VRC07-523LS, in six adults without HIV in part 1 and a multicenter trial of up to six monthly infusions of these three bNAbs in 12 people living with HIV with an antiretroviral therapy (ART) interruption in part 2. The primary endpoints were safety, tolerability and pharmacokinetics, and the secondary endpoints in part 2 were antiviral activity following ART discontinuation, changes in CD4+ T cell counts and development of HIV-1 sequence mutations associated with bNAb resistance. The trial met its prespecified endpoints. The bNAb treatment was generally safe and well tolerated. In part 2, 83% of participants (10 of 12) maintained virologic suppression for the duration of antibody therapy for at least 28 weeks, and 42% of participants (5 of 12) showed virologic suppression for at least 38-44 weeks, despite the decline of serum bNAb concentrations to low or undetectable levels. In exploratory analyses, early viral rebound in two individuals correlated with baseline resistance to PGT121 and PGDM1400, whereas long-term virologic control in five individuals correlated with reduced immune activation, T cell exhaustion and proinflammatory signaling following bNAb therapy. Our data show the potential of a triple bNAb cocktail to suppress HIV-1 in the absence of ART. ClinicalTrials.gov registration: NCT03721510 .
© 2024. The Author(s).
Conflict of interest statement
Competing interests: B.J. is a part-time employee of Leyden Labs, B.V. The other authors declare no competing interests.
Figures














References
Publication types
MeSH terms
Substances
Associated data
Grants and funding
- T32 AI007387/AI/NIAID NIH HHS/United States
- U01 AI145801/AI/NIAID NIH HHS/United States
- AI124377, AI128751, AI145801, AI149670, AI164556, AI169615, AI177687/U.S. Department of Health & Human Services | NIH | National Institute of Allergy and Infectious Diseases (NIAID)
- P01 AI169615/AI/NIAID NIH HHS/United States
- UM1 AI164556/AI/NIAID NIH HHS/United States
- UL1 TR001102/TR/NCATS NIH HHS/United States
- UM1 AI124377/AI/NIAID NIH HHS/United States
- U19 AI128751/AI/NIAID NIH HHS/United States
- K08 AI106408/AI/NIAID NIH HHS/United States
- R01 AI149670/AI/NIAID NIH HHS/United States
- R37 AI150590/AI/NIAID NIH HHS/United States
- K23 AI114381/AI/NIAID NIH HHS/United States
- P01 AI177687/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

