First insight into the genomes of the Pulmonaria officinalis group (Boraginaceae) provided by repeatome analysis and comparative karyotyping
- PMID: 39266954
- PMCID: PMC11395855
- DOI: 10.1186/s12870-024-05497-4
First insight into the genomes of the Pulmonaria officinalis group (Boraginaceae) provided by repeatome analysis and comparative karyotyping
Abstract
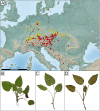
Background: The genus Pulmonaria (Boraginaceae) represents a taxonomically complex group of species in which morphological similarity contrasts with striking karyological variation. The presence of different numbers of chromosomes in the diploid state suggests multiple hybridization/polyploidization events followed by chromosome rearrangements (dysploidy). Unfortunately, the phylogenetic relationships and evolution of the genome, have not yet been elucidated. Our study focused on the P. officinalis group, the most widespread species complex, which includes two morphologically similar species that differ in chromosome number, i.e. P. obscura (2n = 14) and P. officinalis (2n = 16). Ornamental cultivars, morphologically similar to P. officinalis (garden escapes), whose origin is unclear, were also studied. Here, we present a pilot study on genome size and repeatome dynamics of these closely related species in order to gain new information on their genome and chromosome structure.
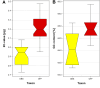
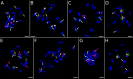
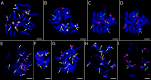
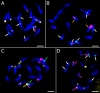
Results: Flow cytometry confirmed a significant difference in genome size between P. obscura and P. officinalis, corresponding to the number of chromosomes. Genome-wide repeatome analysis performed on genome skimming data showed that retrotransposons were the most abundant repeat type, with a higher proportion of Ty3/Gypsy elements, mainly represented by the Tekay lineage. Comparative analysis revealed no species-specific retrotransposons or striking differences in their copy number between the species. A new set of chromosome-specific cytogenetic markers, represented by satellite DNAs, showed that the chromosome structure in P. officinalis was more variable compared to that of P. obscura. Comparative karyotyping supported the hybrid origin of putative hybrids with 2n = 15 collected from a mixed population of both species and outlined the origin of ornamental garden escapes, presumably derived from the P. officinalis complex.
Conclusions: Large-scale genome size analysis and repeatome characterization of the two morphologically similar species of the P. officinalis group improved our knowledge of the genome dynamics and differences in the karyotype structure. A new set of chromosome-specific cytogenetic landmarks was identified and used to reveal the origin of putative hybrids and ornamental cultivars morphologically similar to P. officinalis.
Keywords: Pulmonaria; Comparative karyotyping; Genome size; Repeatome; Satellite DNA.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Chacón J, Luebert F, Hilger HH, Ovchinnikova S, Selvi F, Cecchi L, et al. The borage family (Boraginaceae s.s.): a revised infrafamilial classification based on new phylogenetic evidence, with emphasis on the placement of some enigmatic genera. Taxon. 2016;65(3):523–46. 10.12705/653.6. 10.12705/653.6 - DOI
-
- Merxmüller H, Grau J. Dysploidie Bei Pulmonaria. Rev Roum Biol-Botanique. 1969;14(1):57–63.
-
- Sauer W. Karyo-Systematische Untersuchungen an Der Gattung Pulmonaria (Boraginaceae). Chromosomen-Zahlen, Karyotyp-Analysen und allgemeine Hinweise Auf die Entwicklungsgeschichte. Bibliot Bot. 1975;131:1–85.
-
- Sauer W. A phylogenetic model for North-Eurasiatic Tubifloras on the base of comparative morphology, zytogenetics and ecology. Acta Bot Yunnanica. 1986;8(4):383–95.
-
- Májovský J, Murín A. Cytotaxonomy of the genus Pulmonaria L. in Slovakia. Bol Soc Brot Ser 2. 1980;53:25–739.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

