SIRT1-mediated deacetylation of FOXO3 enhances mitophagy and drives hormone resistance in endometrial cancer
- PMID: 39266959
- PMCID: PMC11391609
- DOI: 10.1186/s10020-024-00915-7
SIRT1-mediated deacetylation of FOXO3 enhances mitophagy and drives hormone resistance in endometrial cancer
Abstract
Background: The complex interplay between Sirtuin 1 (SIRT1) and FOXO3 in endometrial cancer (EC) remains understudied. This research aims to unravel the interactions of deacetylase SIRT1 and transcription factor FOXO3 in EC, focusing on their impact on mitophagy and hormone resistance.
Methods: High-throughput sequencing, cell experiments, and bioinformatics tools were employed to investigate the roles and interactions of SIRT1 and FOXO3 in EC. Co-immunoprecipitation (Co-IP) assay was used to assess the interaction between SIRT1 and FOXO3 in RL95-2 cells. Functional assays were used to assess cell viability, proliferation, migration, invasion, apoptosis, and the expression of related genes and proteins. A mouse model of EC was established to evaluate tumor growth and hormone resistance under different interventions. Immunohistochemistry and TUNEL assays were used to assess protein expression and apoptosis in tumor tissues.
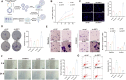
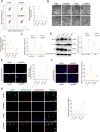
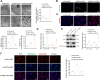
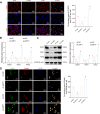
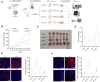
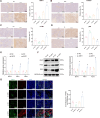
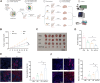
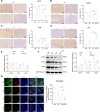
Results: High-throughput transcriptome sequencing revealed a close association between SIRT1, FOXO3, and EC development. Co-IP showed a protein-protein interaction between SIRT1 and FOXO3. Overexpression of SIRT1 enhanced FOXO3 deacetylation and activity, promoting BNIP3 transcription and PINK1/Parkin-mediated mitophagy, which in turn promoted cell proliferation, migration, invasion, and inhibited apoptosis in vitro, as well as increased tumor growth and hormone resistance in vivo. These findings highlighted SIRT1 as an upstream regulator and potential therapeutic target in EC.
Conclusion: This study reveals a novel molecular mechanism underlying the functional relevance of SIRT1 in regulating mitophagy and hormone resistance through the deacetylation of FOXO3 in EC, thereby providing valuable insights for new therapeutic strategies.
Keywords: BNIP3/PINK1/Parkin signaling pathway; Deacetylation modification; Endometrial cancer; FOXO3; Hormone resistance; Mitophagy; SIRT1.
© 2024. The Author(s).
Conflict of interest statement
The author declares no competing interests.
Figures



Similar articles
-
CDK9 inhibition blocks the initiation of PINK1-PRKN-mediated mitophagy by regulating the SIRT1-FOXO3-BNIP3 axis and enhances the therapeutic effects involving mitochondrial dysfunction in hepatocellular carcinoma.Autophagy. 2022 Aug;18(8):1879-1897. doi: 10.1080/15548627.2021.2007027. Epub 2021 Dec 10. Autophagy. 2022. PMID: 34890308 Free PMC article.
-
SIRT1 inhibits chemoresistance and cancer stemness of gastric cancer by initiating an AMPK/FOXO3 positive feedback loop.Cell Death Dis. 2020 Feb 12;11(2):115. doi: 10.1038/s41419-020-2308-4. Cell Death Dis. 2020. PMID: 32051395 Free PMC article.
-
Lapatinib sensitivity in nasopharyngeal carcinoma is modulated by SIRT2-mediated FOXO3 deacetylation.BMC Cancer. 2019 Nov 14;19(1):1106. doi: 10.1186/s12885-019-6308-7. BMC Cancer. 2019. PMID: 31727006 Free PMC article.
-
Sirtuin 1 promotes autophagy and proliferation of endometrial cancer cells by reducing acetylation level of LC3.Cell Biol Int. 2021 May;45(5):1050-1059. doi: 10.1002/cbin.11549. Epub 2021 Feb 2. Cell Biol Int. 2021. PMID: 33438275
-
SIRT1 and gynecological malignancies (Review).Oncol Rep. 2021 Apr;45(4):43. doi: 10.3892/or.2021.7994. Epub 2021 Mar 2. Oncol Rep. 2021. PMID: 33649834 Free PMC article. Review.
Cited by
-
Remimazolam alleviates hepatic ischemia-reperfusion injury by activating FOXO1/3 signaling : Remimazolam alleviates hepatic ischemia reperfusion injury.BMC Gastroenterol. 2025 Apr 22;25(1):283. doi: 10.1186/s12876-025-03820-3. BMC Gastroenterol. 2025. PMID: 40263992 Free PMC article.
-
Cuproptosis-related genes and agents: implications in tumor drug resistance and future perspectives.Front Pharmacol. 2025 May 8;16:1559236. doi: 10.3389/fphar.2025.1559236. eCollection 2025. Front Pharmacol. 2025. PMID: 40406488 Free PMC article. Review.
-
Sirtuins in mitophagy: key gatekeepers of mitochondrial quality.Mol Cell Biochem. 2025 Jul 24. doi: 10.1007/s11010-025-05358-0. Online ahead of print. Mol Cell Biochem. 2025. PMID: 40705152 Review.
-
Mitochondrial metabolism and cancer therapeutic innovation.Signal Transduct Target Ther. 2025 Aug 4;10(1):245. doi: 10.1038/s41392-025-02311-x. Signal Transduct Target Ther. 2025. PMID: 40754534 Free PMC article. Review.
-
CT-Scan-Assessed Body Composition and Its Association with Tumor Protein Expression in Endometrial Cancer: The Role of Muscle and Adiposity Quantities.Cancers (Basel). 2024 Dec 18;16(24):4222. doi: 10.3390/cancers16244222. Cancers (Basel). 2024. PMID: 39766121 Free PMC article.
References
-
- Ashrafizadeh M, Zhang W, Zou R, Sethi G, Klionsky DJ, Zhang X. A bioinformatics analysis, pre-clinical and clinical conception of autophagy in pancreatic cancer: Complexity and simplicity in crosstalk. Pharmacol Res. 2023;194: 106822. 10.1016/j.phrs.2023.106822. 10.1016/j.phrs.2023.106822 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

