Mitogenomes comparison of 3 species of Asparagus L shedding light on their functions due to domestication and adaptative evolution
- PMID: 39266980
- PMCID: PMC11396758
- DOI: 10.1186/s12864-024-10768-3
Mitogenomes comparison of 3 species of Asparagus L shedding light on their functions due to domestication and adaptative evolution
Abstract
Background: Asparagus L., widely distributed in the old world is a genus under Asparagaceae, Asparagales. The species of the genus were mainly used as vegetables, traditional medicines as well as ornamental plants. However, the evolution and functions of mitochondrial (Mt) genomes (mitogenomes) remains largely unknown. In this study, the typical herbal medicine A. taliensis and ornamental plant A. setaceus were used to assemble and annotate the mitogenomes, and the resulting mitogenomes were further compared with published mitogenome of A. officinalis for the analysis of their functions in the context of domestication and adaptative evolution.
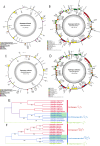
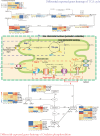
Results: The mitochondrial genomes of both A. taliensis and A. setaceus were assembled as complete circular ones. The phylogenetic trees based on conserved protein-coding genes of Mt genomes and whole chloroplast (Cp) genomes showed that, the phylogenetic relationship of the sampled 13 species of Asparagus L. were not exactly consistent. The collinear analyses between the nuclear (Nu) and Mt genomes confirmed the existence of mutual horizontal genes transfers (HGTs) between Nu and Mt genomes within these species. Based on RNAseq data, the Mt RNA editing were predicted and atp1 and ccmB RNA editing of A. taliensis were further confirmed by DNA sequencing. Simultaneously homologous search found 5 Nu coding gene families including pentatricopeptide-repeats (PPRs) involved in Mt RNA editing. Finally, the Mt genome variations, gene expressions and mutual HGTs between Nu and Mt were detected with correlation to the growth and developmental phenotypes respectively. The results suggest that, both Mt and Nu genomes co-evolved and maintained the Mt organella replication and energy production through TCA and oxidative phosphorylation .
Conclusion: The assembled and annotated complete mitogenomes of both A. taliensis and A. setaceus provide valuable information for their phylogeny and concerted action of Nu and Mt genomes to maintain the energy production system of Asparagus L. in the context of domestication and adaptation to environmental niches.
Keywords: Asparagus L.; Energy production system; Mitochondrial genome; Phylogenetic analysis; RNA editing.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



References
-
- Lei Binbin L, Shuangshuang L, Guozheng W, Yumei S, Aiguo. Hua Jinping: evolutionary analysis of mitochondrial genomes in higher plants. Mol Plant Breed. 2012;10(04):490–500.
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

