Comparative mitochondrial genomics of Terniopsis yongtaiensis in Malpighiales: structural, sequential, and phylogenetic perspectives
- PMID: 39267005
- PMCID: PMC11391645
- DOI: 10.1186/s12864-024-10765-6
Comparative mitochondrial genomics of Terniopsis yongtaiensis in Malpighiales: structural, sequential, and phylogenetic perspectives
Abstract
Background: Terniopsis yongtaiensis, a member of the Podostemaceae family, is an aquatic flowering plant displaying remarkable adaptive traits that enable survival in submerged, turbulent habitats. Despite the progressive expansion of chloroplast genomic information within this family, mitochondrial genome sequences have yet to be reported.
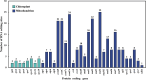
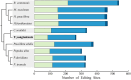
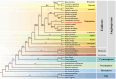
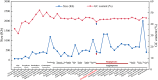
Results: In current study, the mitochondrial genome of the T. yongtaiensis was characterized by a circular genome of 426,928 bp encoding 31 protein-coding genes (PCGs), 18 tRNAs, and 3 rRNA genes. Our comprehensive analysis focused on gene content, repeat sequences, RNA editing processes, intracellular gene transfer, phylogeny, and codon usage bias. Numerous repeat sequences were identified, including 130 simple sequence repeats, 22 tandem repeats, and 220 dispersed repeats. Phylogenetic analysis positioned T. yongtaiensis (Podostemaceae) within the Malpighiales order, showing a close relationship with the Calophyllaceae family, which was consistent with the APG IV classification. A comparative analysis with nine other Malpighiales species revealed both variable and conserved regions, providing insights into the genomic evolution within this order. Notably, the GC content of T. yongtaiensis was distinctively lower compared to other Malpighilales, primarily due to variations in non-coding regions and specific protein-coding genes, particularly the nad genes. Remarkably, the number of RNA editing sites was low (276), distributed unevenly across 27 PCGs. The dN/dS analysis showed only the ccmB gene of T. yongtaiensis was positively selected, which plays a crucial role in cytochrome c biosynthesis. Additionally, there were 13 gene-containing homologous regions between the mitochondrial and chloroplast genomes of T. yongtaiensis, suggesting the gene transfer events between these organellar genomes.
Conclusions: This study assembled and annotated the first mitochondrial genome of the Podostemaceae family. The comparison results of mitochondrial gene composition, GC content, and RNA editing sites provided novel insights into the adaptive traits and genetic reprogramming of this aquatic eudicot group and offered a foundation for future research on the genomic evolution and adaptive mechanisms of Podostemaceae and related plant families in the Malpighiales order.
Keywords: Terniopsis yongtaiensis; Evolution; Genome size variation; Mitochondrial genome; Phylogenetic.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures






Similar articles
-
Assembly and characterization of the first mitochondrial genome of Phyllanthaceae: a case study of the ornamental aquatic plant Phyllanthus fluitans.Genetica. 2025 Jul 16;153(1):25. doi: 10.1007/s10709-025-00241-8. Genetica. 2025. PMID: 40668455
-
Complete mitochondrial genome assembly of Juglans regia unveiled its molecular characteristics, genome evolution, and phylogenetic implications.BMC Genomics. 2024 Sep 28;25(1):894. doi: 10.1186/s12864-024-10818-w. BMC Genomics. 2024. PMID: 39342114 Free PMC article.
-
Insights into structure, codon usage, repeats, and RNA editing of the complete mitochondrial genome of Perilla frutescens (Lamiaceae).Sci Rep. 2024 Jun 17;14(1):13940. doi: 10.1038/s41598-024-64509-3. Sci Rep. 2024. PMID: 38886463 Free PMC article.
-
Characterization and comparative analysis of the complete mitochondrial genome of Phlomoides rotata, a traditional Tibetan medicinal plant.BMC Genomics. 2025 Aug 6;26(1):727. doi: 10.1186/s12864-025-11871-9. BMC Genomics. 2025. PMID: 40770612 Free PMC article.
-
A phylogenetic approach to comparative genomics.Nat Rev Genet. 2025 Jun;26(6):395-405. doi: 10.1038/s41576-024-00803-0. Epub 2025 Jan 8. Nat Rev Genet. 2025. PMID: 39779997 Free PMC article. Review.
Cited by
-
Complete sequencing of the mitochondrial genome of tea plant Camellia sinensis cv. 'Baihaozao': multichromosomal structure, phylogenetic relationships, and adaptive evolutionary analysis.Front Plant Sci. 2025 Jun 13;16:1604404. doi: 10.3389/fpls.2025.1604404. eCollection 2025. Front Plant Sci. 2025. PMID: 40584852 Free PMC article.
-
Assembly and Comparative Analysis of the Complete Mitochondrial Genomes of Smilax glabra and Smilax zeylanica.Genes (Basel). 2025 Apr 14;16(4):450. doi: 10.3390/genes16040450. Genes (Basel). 2025. PMID: 40282410 Free PMC article.
References
-
- Tǎng HT, Kato M. Culture of river-weed Terniopsis chanthaburiensis (Podostemaceae). Aquat Bot. 2020;166:103–255. 10.1016/j.aquabot.2020.103255.10.1016/j.aquabot.2020.103255 - DOI
-
- Rutishauser R. Evolution of unusual morphologies in Lentibulariaceae (bladderworts and allies) and Podostemaceae (river-weeds): a pictorial report at the interface of developmental biology and morphological diversification. Ann Bot. 2016;117:811–32. 10.1093/aob/mcv172. 10.1093/aob/mcv172 - DOI - PMC - PubMed
-
- Taiz L, Zeiger E, Møller IM. Murphy, AS. Plant physiology and development. Sunderland, MA: Sinauer Associates; 2015.
Publication types
MeSH terms
Substances
Grants and funding
- 2020-070705/Special Project of Orchid Survey of National Forestry and Grassland Administration
- 2020-070705/Special Project of Orchid Survey of National Forestry and Grassland Administration
- 2020-070705/Special Project of Orchid Survey of National Forestry and Grassland Administration
- 2020-070705/Special Project of Orchid Survey of National Forestry and Grassland Administration
- 2020-070705/Special Project of Orchid Survey of National Forestry and Grassland Administration
- Grant No.2019-39/National Special Fund for Chinese medicine resources Research in the Public Interest of China
- Grant No.2019-39/National Special Fund for Chinese medicine resources Research in the Public Interest of China
- Grant No.2019-39/National Special Fund for Chinese medicine resources Research in the Public Interest of China
- Grant No.2019-39/National Special Fund for Chinese medicine resources Research in the Public Interest of China
- Grant No.2019-39/National Special Fund for Chinese medicine resources Research in the Public Interest of China
- 2020J05037/Natural Science Foundation of Fujian Province
- JAT190089/Foundation of Fujian Educational Committee
- #32470215/the National Natural Science Foundation of China (NSFC)
LinkOut - more resources
Full Text Sources
Miscellaneous

