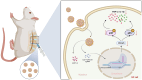
A novel spherical GelMA-HAMA hydrogel encapsulating APET×2 polypeptide and CFIm25-targeting sgRNA for immune microenvironment modulation and nucleus pulposus regeneration in intervertebral discs
- PMID: 39267105
- PMCID: PMC11391743
- DOI: 10.1186/s12951-024-02783-z
A novel spherical GelMA-HAMA hydrogel encapsulating APET×2 polypeptide and CFIm25-targeting sgRNA for immune microenvironment modulation and nucleus pulposus regeneration in intervertebral discs
Abstract
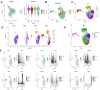
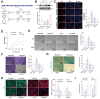
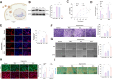
Methods: Single-cell transcriptomics and high-throughput transcriptomics were used to screen factors significantly correlated with intervertebral disc degeneration (IDD). Expression changes of CFIm25 were determined via RT-qPCR and Western blot. NP cells were isolated from mouse intervertebral discs and induced to degrade with TNF-α and IL-1β. CFIm25 was knocked out using CRISPR-Cas9, and CFIm25 knockout and overexpressing nucleus pulposus (NP) cell lines were generated through lentiviral transfection. Proteoglycan expression, protein expression, inflammatory factor expression, cell viability, proliferation, migration, gene expression, and protein expression were analyzed using various assays (alcian blue staining, immunofluorescence, ELISA, CCK-8, EDU labeling, transwell migration, scratch assay, RT-qPCR, Western blot). The GelMA-HAMA hydrogel loaded with APET×2 polypeptide and sgRNA was designed, and its effects on NP regeneration were assessed through in vitro and mouse model experiments. The progression of IDD in mice was evaluated using X-ray, H&E staining, and Safranin O-Fast Green staining. Immunohistochemistry was performed to determine protein expression in NP tissue. Proteomic analysis combined with in vitro and in vivo experiments was conducted to elucidate the mechanisms of hydrogel action.
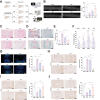
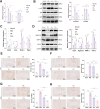
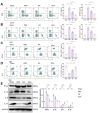
Results: CFIm25 was upregulated in IDD NP tissue and significantly correlated with disease progression. Inhibition of CFIm25 improved NP cell degeneration, enhanced cell proliferation, and migration. The hydrogel effectively knocked down CFIm25 expression, improved NP cell degeneration, promoted cell proliferation and migration, and mitigated IDD progression in a mouse model. The hydrogel inhibited inflammatory factor expression (IL-6, iNOS, IL-1β, TNF-α) by targeting the p38/NF-κB signaling pathway, increased collagen COLII and proteoglycan Aggrecan expression, and suppressed NP degeneration-related factors (COX-2, MMP-3).
Conclusion: The study highlighted the crucial role of CFIm25 in IDD and introduced a promising therapeutic strategy using a porous spherical GelMA-HAMA hydrogel loaded with APET×2 polypeptide and sgRNA. This innovative approach offers new possibilities for treating degenerated intervertebral discs.
Keywords: APET×2 polypeptide; CRISPR-Cas9 protein; Cleavage and polyadenylation specificity factor subunit 5; GelMA-HAMA hydrogel; Immune microenvironment; Intervertebral disc degeneration; Nucleus pulposus regeneration.
© 2024. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



Similar articles
-
Esterase-responsive kartogenin composite hydrogel microspheres boost nucleus pulposus regeneration in intervertebral disc degeneration.Acta Biomater. 2025 May 15;198:131-150. doi: 10.1016/j.actbio.2025.04.001. Epub 2025 Apr 2. Acta Biomater. 2025. PMID: 40180005
-
Injectable hydrogel microspheres promoting inflammation modulation and nucleus pulposus-like differentiation for intervertebral disc regeneration.J Control Release. 2025 Apr 10;380:599-614. doi: 10.1016/j.jconrel.2025.02.016. Epub 2025 Feb 15. J Control Release. 2025. PMID: 39938720
-
Investigating the regulation of the miR-199a-3p/TGF-β/Smad signaling pathway by BSHXF drug-containing serum combined with ADSCs for delaying intervertebral disc degeneration.Front Pharmacol. 2025 Apr 28;16:1583635. doi: 10.3389/fphar.2025.1583635. eCollection 2025. Front Pharmacol. 2025. PMID: 40356987 Free PMC article.
-
[Research progress of hydrogel used for regeneration of nucleus pulposus in intervertebral disc degeneration].Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2020 Mar 15;34(3):275-284. doi: 10.7507/1002-1892.201907092. Zhongguo Xiu Fu Chong Jian Wai Ke Za Zhi. 2020. PMID: 32174070 Free PMC article. Review. Chinese.
-
Differentiation of Pluripotent Stem Cells into Nucleus Pulposus Progenitor Cells for Intervertebral Disc Regeneration.Curr Stem Cell Res Ther. 2019;14(1):57-64. doi: 10.2174/1574888X13666180918095121. Curr Stem Cell Res Ther. 2019. PMID: 30227822 Review.
Cited by
-
Harnessing CRISPR potential for intervertebral disc regeneration strategies.Front Bioeng Biotechnol. 2025 May 8;13:1562412. doi: 10.3389/fbioe.2025.1562412. eCollection 2025. Front Bioeng Biotechnol. 2025. PMID: 40406584 Free PMC article. Review.
References
-
- Yang XX, Yip CH, Zhao S, Ho YP, Chan BP. A bio-inspired nano-material recapitulating the composition, ultra-structure, and function of the glycosaminoglycan-rich extracellular matrix of nucleus pulposus. Biomaterials. 2023;293:121991. 10.1016/j.biomaterials.2022.121991. 10.1016/j.biomaterials.2022.121991 - DOI - PubMed
MeSH terms
Substances
Grants and funding
- 82302763, 82202764, 82202765/National Natural Science Foundation of China
- 82302763, 82202764, 82202765/National Natural Science Foundation of China
- 82302763, 82202764, 82202765/National Natural Science Foundation of China
- 82302763, 82202764, 82202765/National Natural Science Foundation of China
- 82302763, 82202764, 82202765/National Natural Science Foundation of China
- 82302763, 82202764, 82202765/National Natural Science Foundation of China
- 82302763, 82202764, 82202765/National Natural Science Foundation of China
- 82302763, 82202764, 82202765/National Natural Science Foundation of China
- 82302763, 82202764, 82202765/National Natural Science Foundation of China
- 82302763, 82202764, 82202765/National Natural Science Foundation of China
- 82302763, 82202764, 82202765/National Natural Science Foundation of China
- 82302763, 82202764, 82202765/National Natural Science Foundation of China
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

