SEMA6B induces macrophage-mediated inflammation and hepatocyte apoptosis in hepatitis B virus-related acute-on-chronic liver failure
- PMID: 39267780
- PMCID: PMC11388073
- DOI: 10.7150/thno.97007
SEMA6B induces macrophage-mediated inflammation and hepatocyte apoptosis in hepatitis B virus-related acute-on-chronic liver failure
Abstract
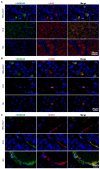
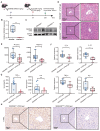
Rationale: Patients with hepatitis B virus-related acute-on-chronic liver failure (HBV-ACLF) have a high short-term mortality rate. Semaphorin-6B (SEMA6B) plays a crucial role in the pathogenesis of HBV-ACLF, but its molecular basis remains unclear. This study aimed to elucidate the mechanisms of SEMA6B in HBV-ACLF progression. Methods: A total of 321 subjects with HBV-ACLF, liver cirrhosis (LC), chronic hepatitis B (CHB), and normal controls (NC) from a prospective multicenter cohort were studied. 84 subjects (HBV-ACLF, n = 50; LC, n = 10; CHB, n = 10; NC, n = 14) among them underwent mRNA sequencing using peripheral blood mononuclear cells (PBMCs) to clarify the mechanisms of SEMA6B in HBV-ACLF. These mechanisms were validated through in vitro studies with hepatocytes and macrophages, as well as in vivo using SEMA6B knockout mice and mice treated with synthetic SEMA6B siRNA. Results: Transcriptome analysis of PBMCs showed that SEMA6B was among the most differentially expressed genes when comparing patients with HBV-ACLF to those with LC, CHB, or NC. ROC analysis demonstrated the reliable diagnostic value of SEMA6B for HBV-ACLF in both the sequencing cohort and an external validation cohort (AUROC = 0.9788 and 0.9026, respectively). SEMA6B levels were significantly higher in the HBV-ACLF patients, especially in non-survivors, with high expression mainly observed in macrophages and hepatocytes in liver tissue. Genes significantly associated with highly expressed SEMA6B were enriched in inflammation and apoptosis pathways in HBV-ACLF non-survivors. Overexpression of SEMA6B in macrophages activated systemic inflammatory responses, while its overexpression in hepatocytes inhibited proliferation through G0/G1 cell cycle arrest and induced apoptosis. Knocking out SEMA6B rescued mice with liver failure by improving liver functions, reducing inflammatory responses, and decreasing hepatocyte apoptosis. Transcriptome analysis of liver tissue showed that SEMA6B knockout significantly ameliorated the liver failure signature, significantly downregulating inflammation-related pathways. Importantly, therapeutic delivery of synthetic SEMA6B siRNA also improved liver function, and reduced both inflammation and hepatocyte apoptosis in mice with liver failure. Conclusion: SEMA6B, a potential diagnostic biomarker for HBV-ACLF, exacerbates liver failure through macrophage-mediated systemic inflammation and hepatocyte apoptosis. These findings highlight SEMA6B as a promising early treatment target for HBV-ACLF patients.
Keywords: acute-on-chronic liver failure; apoptosis; biomarker; hepatitis B virus; inflammation.
© The author(s).
Conflict of interest statement
Competing Interests: The authors have declared that no competing interest exists.
Figures







References
-
- Bernal W, Jalan R, Quaglia A, Simpson K, Wendon J, Burroughs A. Acute-on-chronic liver failure. Lancet. 2015;386:1576–87. - PubMed
-
- Wu T, Li J, Shao L, Xin J, Jiang L, Zhou Q. et al. Development of diagnostic criteria and a prognostic score for hepatitis B virus-related acute-on-chronic liver failure. Gut. 2018;67:2181–91. - PubMed
-
- Li J, Liang X, You S, Feng T, Zhou X, Zhu B. et al. Development and validation of a new prognostic score for hepatitis B virus-related acute-on-chronic liver failure. J Hepatol. 2021;75:1104–15. - PubMed
-
- Arroyo V, Angeli P, Moreau R, Jalan R, Clària J, Trebicka J. et al. The systemic inflammation hypothesis: Towards a new paradigm of acute decompensation and multiorgan failure in cirrhosis. J Hepatol. 2021;74:670–85. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

